High variability of mitochondrial gene order among fungi
- PMID: 24504088
- PMCID: PMC3942027
- DOI: 10.1093/gbe/evu028
High variability of mitochondrial gene order among fungi
Abstract
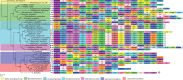
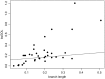
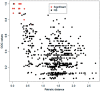
From their origin as an early alpha proteobacterial endosymbiont to their current state as cellular organelles, large-scale genomic reorganization has taken place in the mitochondria of all main eukaryotic lineages. So far, most studies have focused on plant and animal mitochondrial (mt) genomes (mtDNA), but fungi provide new opportunities to study highly differentiated mtDNAs. Here, we analyzed 38 complete fungal mt genomes to investigate the evolution of mtDNA gene order among fungi. In particular, we looked for evidence of nonhomologous intrachromosomal recombination and investigated the dynamics of gene rearrangements. We investigated the effect that introns, intronic open reading frames (ORFs), and repeats may have on gene order. Additionally, we asked whether the distribution of transfer RNAs (tRNAs) evolves independently to that of mt protein-coding genes. We found that fungal mt genomes display remarkable variation between and within the major fungal phyla in terms of gene order, genome size, composition of intergenic regions, and presence of repeats, introns, and associated ORFs. Our results support previous evidence for the presence of mt recombination in all fungal phyla, a process conspicuously lacking in most Metazoa. Overall, the patterns of rearrangements may be explained by the combined influences of recombination (i.e., most likely nonhomologous and intrachromosomal), accumulated repeats, especially at intergenic regions, and to a lesser extent, mobile element dynamics.
Keywords: Basidiomycota; basal fungi; endosymbiosis; fungal phylogeny; genome size reduction; rearrangement rates; sordariomycetes.
Figures

References
-
- Adams KL, Palmer JD. Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenet Evol. 2003;29:380–395. - PubMed
-
- Al-Reedy RM, Malireddy R, Dillman CB, Kennell JC. Comparative analysis of Fusarium mitochondrial genomes reveals a highly variable region that encodes an exceptionally large open reading frame. Fungal Genet Biol. 2012;49:2–14. - PubMed
-
- Amlacher S, et al. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell. 2011;146:277–289. - PubMed
-
- Barr CM, Neiman M, Taylor DR. Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol. 2005;168:39–50. - PubMed
-
- Bartelli TF, Ferreira RC, Colombo AL, Briones MRS. Intraspecific comparative genomics of Candida albicans mitochondria reveals non-coding regions under neutral evolution. Infect Genet Evol. 2013;14:302–312. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

