Thioredoxin targets fundamental processes in a methane-producing archaeon, Methanocaldococcus jannaschii
- PMID: 24505058
- PMCID: PMC3932849
- DOI: 10.1073/pnas.1324240111
Thioredoxin targets fundamental processes in a methane-producing archaeon, Methanocaldococcus jannaschii
Abstract
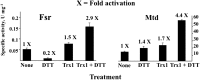
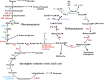
Thioredoxin (Trx), a small redox protein, controls multiple processes in eukaryotes and bacteria by changing the thiol redox status of selected proteins. The function of Trx in archaea is, however, unexplored. To help fill this gap, we have investigated this aspect in methanarchaea--strict anaerobes that produce methane, a fuel and greenhouse gas. Bioinformatic analyses suggested that Trx is nearly universal in methanogens. Ancient methanogens that produce methane almost exclusively from H2 plus CO2 carried approximately two Trx homologs, whereas nutritionally versatile members possessed four to eight. Due to its simplicity, we studied the Trx system of Methanocaldococcus jannaschii--a deeply rooted hyperthermophilic methanogen growing only on H2 plus CO2. The organism carried two Trx homologs, canonical Trx1 that reduced insulin and accepted electrons from Escherichia coli thioredoxin reductase and atypical Trx2. Proteomic analyses with air-oxidized extracts treated with reduced Trx1 revealed 152 potential targets representing a range of processes--including methanogenesis, biosynthesis, transcription, translation, and oxidative response. In enzyme assays, Trx1 activated two selected targets following partial deactivation by O2, validating proteomics observations: methylenetetrahydromethanopterin dehydrogenase, a methanogenesis enzyme, and sulfite reductase, a detoxification enzyme. The results suggest that Trx assists methanogens in combating oxidative stress and synchronizing metabolic activities with availability of reductant, making it a critical factor in the global carbon cycle and methane emission. Because methanogenesis developed before the oxygenation of Earth, it seems possible that Trx functioned originally in metabolic regulation independently of O2, thus raising the question whether a complex biological system of this type evolved at least 2.5 billion years ago.
Keywords: early Earth; evolution; hydrothermal vent; methanogenic archaea; redox regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Meyer Y, Buchanan BB, Vignols F, Reichheld JP. Thioredoxins and glutaredoxins: Unifying elements in redox biology. Annu Rev Genet. 2009;43:335–367. - PubMed
-
- Jeon SJ, Ishikawa K. Identification and characterization of thioredoxin and thioredoxin reductase from Aeropyrum pernix K1. Eur J Biochem. 2002;269(22):5423–5430. - PubMed
-
- Kashima Y, Ishikawa K. A hyperthermostable novel protein-disulfide oxidoreductase is reduced by thioredoxin reductase from hyperthermophilic archaeon Pyrococcus horikoshii. Arch Biochem Biophys. 2003;418(2):179–185. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

