Whole-genome array CGH evaluation for replacing prenatal karyotyping in Hong Kong
- PMID: 24505343
- PMCID: PMC3914896
- DOI: 10.1371/journal.pone.0087988
Whole-genome array CGH evaluation for replacing prenatal karyotyping in Hong Kong
Abstract
Objective: To evaluate the effectiveness of whole-genome array comparative genomic hybridization (aCGH) in prenatal diagnosis in Hong Kong.
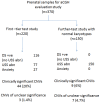
Methods: Array CGH was performed on 220 samples recruited prospectively as the first-tier test study. In addition 150 prenatal samples with abnormal fetal ultrasound findings found to have normal karyotypes were analyzed as a 'further-test' study using NimbleGen CGX-135K oligonucleotide arrays.
Results: Array CGH findings were concordant with conventional cytogenetic results with the exception of one case of triploidy. It was found in the first-tier test study that aCGH detected 20% (44/220) clinically significant copy number variants (CNV), of which 21 were common aneuploidies and 23 had other chromosomal imbalances. There were 3.2% (7/220) samples with CNVs detected by aCGH but not by conventional cytogenetics. In the 'further-test' study, the additional diagnostic yield of detecting chromosome imbalance was 6% (9/150). The overall detection for CNVs of unclear clinical significance was 2.7% (10/370) with 0.9% found to be de novo. Eleven loci of common CNVs were found in the local population.
Conclusion: Whole-genome aCGH offered a higher resolution diagnostic capacity than conventional karyotyping for prenatal diagnosis either as a first-tier test or as a 'further-test' for pregnancies with fetal ultrasound anomalies. We propose replacing conventional cytogenetics with aCGH for all pregnancies undergoing invasive diagnostic procedures after excluding common aneuploidies and triploidies by quantitative fluorescent PCR. Conventional cytogenetics can be reserved for visualization of clinically significant CNVs.
Conflict of interest statement
Figures


References
-
- Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, et al. (2010) Consensus statement: Chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet 86: 749–764 10.1016/j.ajhg.2010.04.006 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

