Regulation of immunoglobulin class-switch recombination: choreography of noncoding transcription, targeted DNA deamination, and long-range DNA repair
- PMID: 24507154
- PMCID: PMC4150736
- DOI: 10.1016/B978-0-12-800267-4.00001-8
Regulation of immunoglobulin class-switch recombination: choreography of noncoding transcription, targeted DNA deamination, and long-range DNA repair
Abstract
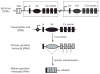
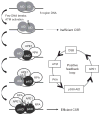
Upon encountering antigens, mature IgM-positive B lymphocytes undergo class-switch recombination (CSR) wherein exons encoding the default Cμ constant coding gene segment of the immunoglobulin (Ig) heavy-chain (Igh) locus are excised and replaced with a new constant gene segment (referred to as "Ch genes", e.g., Cγ, Cɛ, or Cα). The B cell thereby changes from expressing IgM to one producing IgG, IgE, or IgA, with each antibody isotype having a different effector function during an immune reaction. CSR is a DNA deletional-recombination reaction that proceeds through the generation of DNA double-strand breaks (DSBs) in repetitive switch (S) sequences preceding each Ch gene and is completed by end-joining between donor Sμ and acceptor S regions. CSR is a multistep reaction requiring transcription through S regions, the DNA cytidine deaminase AID, and the participation of several general DNA repair pathways including base excision repair, mismatch repair, and classical nonhomologous end-joining. In this review, we discuss our current understanding of how transcription through S regions generates substrates for AID-mediated deamination and how AID participates not only in the initiation of CSR but also in the conversion of deaminated residues into DSBs. Additionally, we review the multiple processes that regulate AID expression and facilitate its recruitment specifically to the Ig loci, and how deregulation of AID specificity leads to oncogenic translocations. Finally, we summarize recent data on the potential role of AID in the maintenance of the pluripotent stem cell state during epigenetic reprogramming.
Keywords: Activation-induced deaminase; Class-switch recombination; Induced pluripotency; Lymphomagenesis.
© 2014 Elsevier Inc. All rights reserved.
Figures

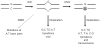





References
-
- Alt FW, Rosenberg N, Casanova RJ, Thomas E, Baltimore D. Immunoglobulin heavy-chain expression and class switching in a murine leukaemia cell line. Nature. 1982;296(5855):325–331. - PubMed
-
- Bardwell PD, Woo CJ, Wei K, Li Z, Martin A, Sack SZ, et al. Altered somatic hypermutation and reduced class-switch recombination in exonuclease 1-mutant mice. Nature Immunology. 2004;5(2):224–229. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

