Engrailed2 modulates cerebellar granule neuron precursor proliferation, differentiation and insulin-like growth factor 1 signaling during postnatal development
- PMID: 24507165
- PMCID: PMC3932947
- DOI: 10.1186/2040-2392-5-9
Engrailed2 modulates cerebellar granule neuron precursor proliferation, differentiation and insulin-like growth factor 1 signaling during postnatal development
Abstract
Background: The homeobox transcription factor Engrailed2 (En2) has been studied extensively in neurodevelopment, particularly in the midbrain/hindbrain region and cerebellum, where it exhibits dynamic patterns of expression and regulates cell patterning and morphogenesis. Because of its roles in regulating cerebellar development and evidence of cerebellar pathology in autism spectrum disorder (ASD), we previously examined an ENGRAILED2 association and found evidence to support EN2 as a susceptibility gene, a finding replicated by several other investigators. However, its functions at the cell biological level remain undefined. In the mouse, En2 gene is expressed in granule neuron precursors (GNPs) just as they exit the cell cycle and begin to differentiate, raising the possibility that En2 may modulate these developmental processes.
Methods: To define En2 functions, we examined proliferation, differentiation and signaling pathway activation in En2 knockout (KO) and wild-type (WT) GNPs in response to a variety of extracellular growth factors and following En2 cDNA overexpression in cell culture. In vivo analyses of cerebellar GNP proliferation as well as responses to insulin-like growth factor-1 (IGF1) treatment were also conducted.
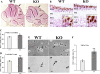
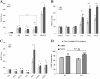
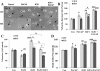
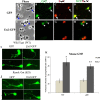
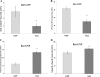
Results: Proliferation markers were increased in KO GNPs in vivo and in 24-h cultures, suggesting En2 normally serves to promote cell cycle exit. Significantly, IGF1 stimulated greater DNA synthesis in KO than WT cells in culture, a finding associated with markedly increased phospho-S6 kinase activation. Similarly, there was three-fold greater DNA synthesis in the KO cerebellum in response to IGF1 in vivo. On the other hand, KO GNPs exhibited reduced neurite outgrowth and differentiation. Conversely, En2 overexpression increased cell cycle exit and promoted neuronal differentiation.
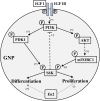
Conclusions: In aggregate, our observations suggest that the ASD-associated gene En2 promotes GNP cell cycle exit and differentiation, and modulates IGF1 activity during postnatal cerebellar development. Thus, genetic/epigenetic alterations of EN2 expression may impact proliferation, differentiation and IGF1 signaling as possible mechanisms that may contribute to ASD pathogenesis.
Figures

Similar articles
-
The Transcriptional Regulator SnoN Promotes the Proliferation of Cerebellar Granule Neuron Precursors in the Postnatal Mouse Brain.J Neurosci. 2019 Jan 2;39(1):44-62. doi: 10.1523/JNEUROSCI.0688-18.2018. Epub 2018 Nov 13. J Neurosci. 2019. PMID: 30425119 Free PMC article.
-
Engrailed-2 is a cell autonomous regulator of neurogenesis in cultured hippocampal neural stem cells.Dev Neurobiol. 2021 Jul;81(5):724-735. doi: 10.1002/dneu.22824. Epub 2021 May 5. Dev Neurobiol. 2021. PMID: 33852756
-
Autism associated gene, engrailed2, and flanking gene levels are altered in post-mortem cerebellum.PLoS One. 2014 Feb 10;9(2):e87208. doi: 10.1371/journal.pone.0087208. eCollection 2014. PLoS One. 2014. PMID: 24520327 Free PMC article.
-
Engrailed-2 (En2) deletion produces multiple neurodevelopmental defects in monoamine systems, forebrain structures and neurogenesis and behavior.Hum Mol Genet. 2015 Oct 15;24(20):5805-27. doi: 10.1093/hmg/ddv301. Epub 2015 Jul 28. Hum Mol Genet. 2015. PMID: 26220976 Free PMC article.
-
Two Pax2/5/8-binding sites in Engrailed2 are required for proper initiation of endogenous mid-hindbrain expression.Mech Dev. 2000 Feb;90(2):155-65. doi: 10.1016/s0925-4773(99)00242-7. Mech Dev. 2000. PMID: 10640701
Cited by
-
The Cerebellum-Ventral Tegmental Area Microcircuit and Its Implications for Autism Spectrum Disorder: A Narrative Review.Neuropsychiatr Dis Treat. 2024 Oct 29;20:2039-2048. doi: 10.2147/NDT.S485487. eCollection 2024. Neuropsychiatr Dis Treat. 2024. PMID: 39494383 Free PMC article. Review.
-
Developmental Role of Adenosine Kinase in the Cerebellum.eNeuro. 2021 Jun 2;8(3):ENEURO.0011-21.2021. doi: 10.1523/ENEURO.0011-21.2021. Print 2021 May-Jun. eNeuro. 2021. PMID: 33863781 Free PMC article.
-
Rapid Detection of Neurodevelopmental Phenotypes in Human Neural Precursor Cells (NPCs).J Vis Exp. 2018 Mar 2;(133):56628. doi: 10.3791/56628. J Vis Exp. 2018. PMID: 29553565 Free PMC article.
-
GH Dysfunction in Engrailed-2 Knockout Mice, a Model for Autism Spectrum Disorders.Front Pediatr. 2014 Sep 1;2:92. doi: 10.3389/fped.2014.00092. eCollection 2014. Front Pediatr. 2014. PMID: 25225635 Free PMC article.
-
Dysregulation of mTOR signaling mediates common neurite and migration defects in both idiopathic and 16p11.2 deletion autism neural precursor cells.Elife. 2024 Mar 25;13:e82809. doi: 10.7554/eLife.82809. Elife. 2024. PMID: 38525876 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

