The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria
- PMID: 24508234
- PMCID: PMC3964146
- DOI: 10.1016/j.immuni.2014.01.003
The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria
Abstract
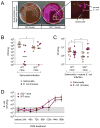
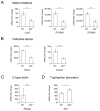
Interleukin-22 (IL-22) is highly induced in response to infections with a variety of pathogens, and its main functions are considered to be tissue repair and host defense at mucosal surfaces. Here we showed that IL-22 has a unique role during infection in that its expression suppressed the intestinal microbiota and enhanced the colonization of a pathogen. IL-22 induced the expression of antimicrobial proteins, including lipocalin-2 and calprotectin, which sequester essential metal ions from microbes. Because Salmonella enterica ser. Typhimurium can overcome metal ion starvation mediated by lipocalin-2 and calprotectin via alternative pathways, IL-22 boosted its colonization of the inflamed intestine by suppressing commensal Enterobacteriaceae, which are susceptible to the antimicrobial proteins. Thus, IL-22 tipped the balance between pathogenic and commensal bacteria in favor of a pathogen. Taken together, IL-22 induction can be exploited by pathogens to suppress the growth of their closest competitors, thereby enhancing pathogen colonization of mucosal surfaces.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures





Comment in
-
The battle in the gut.Immunity. 2014 Feb 20;40(2):173-5. doi: 10.1016/j.immuni.2014.01.007. Immunity. 2014. PMID: 24560194
References
-
- Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Russmann H, Hardt WD. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infection and immunity. 2003;71:2839–2858. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

