The dsRBP and inactive editor ADR-1 utilizes dsRNA binding to regulate A-to-I RNA editing across the C. elegans transcriptome
- PMID: 24508457
- PMCID: PMC3959997
- DOI: 10.1016/j.celrep.2014.01.011
The dsRBP and inactive editor ADR-1 utilizes dsRNA binding to regulate A-to-I RNA editing across the C. elegans transcriptome
Abstract
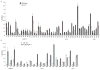
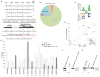
Inadequate adenosine-to-inosine editing of noncoding regions occurs in disease but is often uncorrelated with ADAR levels, underscoring the need to study deaminase-independent control of editing. C. elegans have two ADAR proteins, ADR-2 and the theoretically catalytically inactive ADR-1. Using high-throughput RNA sequencing of wild-type and adr mutant worms, we expand the repertoire of C. elegans edited transcripts over 5-fold and confirm that ADR-2 is the only active deaminase in vivo. Despite lacking deaminase function, ADR-1 affects editing of over 60 adenosines within the 3' UTRs of 16 different mRNAs. Furthermore, ADR-1 interacts directly with ADR-2 substrates, even in the absence of ADR-2, and mutations within its double-stranded RNA (dsRNA) binding domains abolish both binding and editing regulation. We conclude that ADR-1 acts as a major regulator of editing by binding ADR-2 substrates in vivo. These results raise the possibility that other dsRNA binding proteins, including the inactive human ADARs, regulate RNA editing through deaminase-independent mechanisms.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

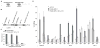
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

