E3 ubiquitin ligase Cbl-b suppresses proallergic T cell development and allergic airway inflammation
- PMID: 24508458
- PMCID: PMC3969736
- DOI: 10.1016/j.celrep.2014.01.012
E3 ubiquitin ligase Cbl-b suppresses proallergic T cell development and allergic airway inflammation
Abstract
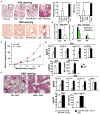
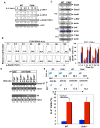
E3 ubiquitin ligase Cbl-b has emerged as a gatekeeper that controls the activation threshold of the T cell antigen receptor and maintains the balance between tolerance and autoimmunity. Here, we report that the loss of Cbl-b facilitates T helper 2 (Th2) and Th9 cell differentiation in vitro. In a mouse model of asthma, the absence of Cbl-b results in severe airway inflammation and stronger Th2 and Th9 responses. Mechanistically, Cbl-b selectively associates with Stat6 upon IL-4 ligation and targets Stat6 for ubiquitination and degradation. These processes are heightened in the presence of T cell receptor (TCR)/CD28 costimulation. Furthermore, we identify K108 and K398 as Stat6 ubiquitination sites. Intriguingly, introducing Stat6 deficiency into Cblb(-/-) mice abrogates hyper-Th2 responses but only partially attenuates Th9 responses. Therefore, our data reveal a function for Cbl-b in the regulation of Th2 and Th9 cell differentiation.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





Similar articles
-
c-CBL/LCK/c-JUN/ETS1/CD28 axis restrains childhood asthma by suppressing Th2 differentiation.Mol Med. 2024 Sep 28;30(1):164. doi: 10.1186/s10020-024-00872-1. Mol Med. 2024. PMID: 39342146 Free PMC article.
-
E3 ubiquitin ligase Cbl-b regulates Pten via Nedd4 in T cells independently of its ubiquitin ligase activity.Cell Rep. 2012 May 31;1(5):472-82. doi: 10.1016/j.celrep.2012.04.008. Cell Rep. 2012. PMID: 22763434 Free PMC article.
-
Protein Tyrosine Phosphatase SHP-1 Modulates T Cell Responses by Controlling Cbl-b Degradation.J Immunol. 2015 Nov 1;195(9):4218-27. doi: 10.4049/jimmunol.1501200. Epub 2015 Sep 28. J Immunol. 2015. PMID: 26416283 Free PMC article.
-
Ubiquitin ligase Cbl-b and inhibitory Cblin peptides.Biochim Biophys Acta Proteins Proteom. 2020 Nov;1868(11):140495. doi: 10.1016/j.bbapap.2020.140495. Epub 2020 Jul 12. Biochim Biophys Acta Proteins Proteom. 2020. PMID: 32663526 Review.
-
Regulation of peripheral T cell tolerance by the E3 ubiquitin ligase Cbl-b.Semin Immunol. 2007 Jun;19(3):206-14. doi: 10.1016/j.smim.2007.02.004. Epub 2007 Mar 27. Semin Immunol. 2007. PMID: 17391982 Review.
Cited by
-
Posttranscriptional regulation of T helper cell fate decisions.J Cell Biol. 2018 Aug 6;217(8):2615-2631. doi: 10.1083/jcb.201708075. Epub 2018 Apr 23. J Cell Biol. 2018. PMID: 29685903 Free PMC article. Review.
-
Solitary fibrous tumors: loss of chimeric protein expression and genomic instability mark dedifferentiation.Mod Pathol. 2015 Aug;28(8):1074-83. doi: 10.1038/modpathol.2015.70. Epub 2015 May 29. Mod Pathol. 2015. PMID: 26022454
-
STAT6 degradation and ubiquitylated TRIML2 are essential for activation of human oncogenic herpesvirus.PLoS Pathog. 2018 Dec 10;14(12):e1007416. doi: 10.1371/journal.ppat.1007416. eCollection 2018 Dec. PLoS Pathog. 2018. PMID: 30532138 Free PMC article.
-
CD28 and ICOS in immune regulation: Structural insights and therapeutic targeting.Bioorg Med Chem Lett. 2025 Nov 1;127:130310. doi: 10.1016/j.bmcl.2025.130310. Epub 2025 Jun 15. Bioorg Med Chem Lett. 2025. PMID: 40527414 Review.
-
Ubiquitination regulates allergic asthma by affecting immune cells and immune responses.Biochem Biophys Rep. 2025 Aug 19;43:102212. doi: 10.1016/j.bbrep.2025.102212. eCollection 2025 Sep. Biochem Biophys Rep. 2025. PMID: 40893770 Free PMC article. Review.
References
-
- Bachmaier K, Krawczyk C, Kozieradzki I, Kong YY, Sasaki T, Oliveira-dos-Santos AJ, Mariathasan S, Bouchard D, Wakeham A, Itie A, et al. Negative regulation of lymphocyte activation and autoimmunity by the molecular adaptor Cbl-b. Nature. 2000;403:211–216. - PubMed
-
- Bachmaier K, Toya S, Gao X, Triantafillou T, Garrean S, Park GY, Frey RS, Vogel S, Minshall R, Christman JW, et al. E3 ubiquitin ligase Cblb regulates the acute inflammatory response underlying lung injury. Nat Med. 2007;13:920–926. - PubMed
-
- Bettelli E, Oukka M, Kuchroo VK. T(H)-17 cells in the circle of immunity and autoimmunity. Nat Immunol. 2007;8:345–350. - PubMed
-
- Chatila TA. Interleukin-4 receptor signaling pathways in asthma pathogenesis. Trends Mol Med. 2004;10:493–499. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

