The mthA mutation conferring low-level resistance to streptomycin enhances antibiotic production in Bacillus subtilis by increasing the S-adenosylmethionine pool size
- PMID: 24509311
- PMCID: PMC3993368
- DOI: 10.1128/JB.01441-13
The mthA mutation conferring low-level resistance to streptomycin enhances antibiotic production in Bacillus subtilis by increasing the S-adenosylmethionine pool size
Abstract
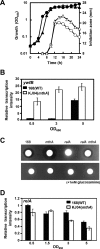
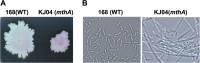
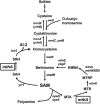
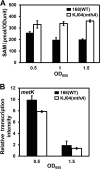
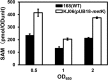
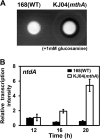
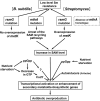
Certain Str(r) mutations that confer low-level streptomycin resistance result in the overproduction of antibiotics by Bacillus subtilis. Using comparative genome-sequencing analysis, we successfully identified this novel mutation in B. subtilis as being located in the mthA gene, which encodes S-adenosylhomocysteine/methylthioadenosine nucleosidase, an enzyme involved in the S-adenosylmethionine (SAM)-recycling pathways. Transformation experiments showed that this mthA mutation was responsible for the acquisition of low-level streptomycin resistance and overproduction of bacilysin. The mthA mutant had an elevated level of intracellular SAM, apparently acquired by arresting SAM-recycling pathways. This increase in the SAM level was directly responsible for bacilysin overproduction, as confirmed by forced expression of the metK gene encoding SAM synthetase. The mthA mutation fully exerted its effect on antibiotic overproduction in the genetic background of rel(+) but not the rel mutant, as demonstrated using an mthA relA double mutant. Strikingly, the mthA mutation activated, at the transcription level, even the dormant ability to produce another antibiotic, neotrehalosadiamine, at concentrations of 150 to 200 μg/ml, an antibiotic not produced (<1 μg/ml) by the wild-type strain. These findings establish the significance of SAM in initiating bacterial secondary metabolism. They also suggest a feasible methodology to enhance or activate antibiotic production, by introducing either the rsmG mutation to Streptomyces or the mthA mutation to eubacteria, since many eubacteria have mthA homologues.
Figures

References
-
- Schatz A, Waksman SA. 1944. Effect of streptomycin and other antibiotic substances upon Mycobacterium tuberculosis and related organisms. Proc. Soc. Exp. Bio. Med. 57:244–248. 10.3181/00379727-57-14769 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

