Dense genomic sampling identifies highways of pneumococcal recombination
- PMID: 24509479
- PMCID: PMC3970364
- DOI: 10.1038/ng.2895
Dense genomic sampling identifies highways of pneumococcal recombination
Abstract
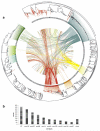
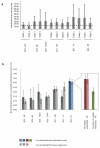
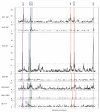
Evasion of clinical interventions by Streptococcus pneumoniae occurs through selection of non-susceptible genomic variants. We report whole-genome sequencing of 3,085 pneumococcal carriage isolates from a 2.4-km(2) refugee camp. This sequencing provides unprecedented resolution of the process of recombination and its impact on population evolution. Genomic recombination hotspots show remarkable consistency between lineages, indicating common selective pressures acting at certain loci, particularly those associated with antibiotic resistance. Temporal changes in antibiotic consumption are reflected in changes in recombination trends, demonstrating rapid spread of resistance when selective pressure is high. The highest frequencies of receipt and donation of recombined DNA fragments were observed in non-encapsulated lineages, implying that this largely overlooked pneumococcal group, which is beyond the reach of current vaccines, may have a major role in genetic exchange and the adaptation of the species as a whole. These findings advance understanding of pneumococcal population dynamics and provide information for the design of future intervention strategies.
Figures

References
-
- O’Brien KL, Nohynek H. Report from a WHO Working Group: standard method for detecting upper respiratory carriage of Streptococcus pneumoniae. The Pediatric infectious disease journal. 2003;22:e1–11. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

