Stage-specific control of early B cell development by the transcription factor Ikaros
- PMID: 24509509
- PMCID: PMC5790181
- DOI: 10.1038/ni.2828
Stage-specific control of early B cell development by the transcription factor Ikaros
Abstract
The transcription factor Ikaros is an essential regulator of lymphopoiesis. Here we studied its B cell-specific function by conditional inactivation of the gene encoding Ikaros (Ikzf1) in pro-B cells. B cell development was arrested at an aberrant 'pro-B cell' stage characterized by increased cell adhesion and loss of signaling via the pre-B cell signaling complex (pre-BCR). Ikaros activated genes encoding signal transducers of the pre-BCR and repressed genes involved in the downregulation of pre-BCR signaling and upregulation of the integrin signaling pathway. Unexpectedly, derepression of expression of the transcription factor Aiolos did not compensate for the loss of Ikaros in pro-B cells. Ikaros induced or suppressed active chromatin at regulatory elements of activated or repressed target genes. Notably, binding of Ikaros and expression of its target genes were dynamically regulated at distinct stages of early B lymphopoiesis.
Figures
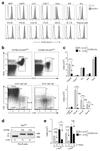
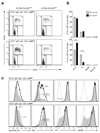



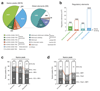
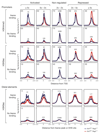

References
-
- Georgopoulos K. Haematopoietic cell-fate decisions, chromatin regulation and Ikaros. Nature Rev Immunol. 2002;2:162–174. - PubMed
-
- Wang J-H, et al. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–549. - PubMed
-
- Mullighan CG, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–114. - PubMed
Online Methods References
-
- Wang J-H, et al. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–549. - PubMed
-
- Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–267. - PubMed
-
- Driegen S, et al. A generic tool for biotinylation of tagged proteins in transgenic mice. Transgenic Res. 2005;14:477–482. - PubMed
-
- Shinkai Y, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

