Lactose-inducible system for metabolic engineering of Clostridium ljungdahlii
- PMID: 24509933
- PMCID: PMC3993169
- DOI: 10.1128/AEM.03666-13
Lactose-inducible system for metabolic engineering of Clostridium ljungdahlii
Abstract
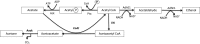
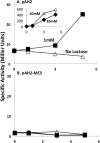
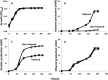
The development of tools for genetic manipulation of Clostridium ljungdahlii has increased its attractiveness as a chassis for autotrophic production of organic commodities and biofuels from syngas and microbial electrosynthesis and established it as a model organism for the study of the basic physiology of acetogenesis. In an attempt to expand the genetic toolbox for C. ljungdahlii, the possibility of adapting a lactose-inducible system for gene expression, previously reported for Clostridium perfringens, was investigated. The plasmid pAH2, originally developed for C. perfringens with a gusA reporter gene, functioned as an effective lactose-inducible system in C. ljungdahlii. Lactose induction of C. ljungdahlii containing pB1, in which the gene for the aldehyde/alcohol dehydrogenase AdhE1 was downstream of the lactose-inducible promoter, increased expression of adhE1 30-fold over the wild-type level, increasing ethanol production 1.5-fold, with a corresponding decrease in acetate production. Lactose-inducible expression of adhE1 in a strain in which adhE1 and the adhE1 homolog adhE2 had been deleted from the chromosome restored ethanol production to levels comparable to those in the wild-type strain. Inducing expression of adhE2 similarly failed to restore ethanol production, suggesting that adhE1 is the homolog responsible for ethanol production. Lactose-inducible expression of the four heterologous genes necessary to convert acetyl coenzyme A (acetyl-CoA) to acetone diverted ca. 60% of carbon flow to acetone production during growth on fructose, and 25% of carbon flow went to acetone when carbon monoxide was the electron donor. These studies demonstrate that the lactose-inducible system described here will be useful for redirecting carbon and electron flow for the biosynthesis of products more valuable than acetate. Furthermore, this tool should aid in optimizing microbial electrosynthesis and for basic studies on the physiology of acetogenesis.
Figures

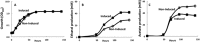


References
-
- Cotter JL, Chinn MS, Grunden AM,. 2009. Influence of process parameters on growth of Clostridium ljungdahlii and Clostridium autoethanogenum on synthesis gas. Enzyme Microb. Technol. 44:281–288. 10.1016/j.enzmictec.2008.11.002 - DOI
-
- Tirado-Acevedo O, Cotter JL, Chinn MS, Grunden AM. 2011. Influence of carbon source pre-adapatation on Clostridium ljungdahlii growth and product formation. J. Bioprocess. Biotech. S2:001
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

