Production of shikimic acid from Escherichia coli through chemically inducible chromosomal evolution and cofactor metabolic engineering
- PMID: 24512078
- PMCID: PMC3923554
- DOI: 10.1186/1475-2859-13-21
Production of shikimic acid from Escherichia coli through chemically inducible chromosomal evolution and cofactor metabolic engineering
Abstract
Background: Shikimic acid (SA) produced from the seeds of Chinese star anise (Illicium verum) is a key intermediate for the synthesis of neuraminidase inhibitors such as oseltamivir (Tamiflu®), an anti-influenza drug. However, plants cannot deliver a stable supply of SA. To avoid the resulting shortages and price fluctuations, a stable source of affordable SA is required. Although recent achievements in metabolic engineering of Escherichia coli strains have significantly increased SA productivity, commonly-used plasmid-based expression systems are prone to genetic instability and require constant selective pressure to ensure plasmid maintenance. Cofactors also play an important role in the biosynthesis of different fermentation products. In this study, we first constructed an E. coli SA production strain that carries no plasmid or antibiotic marker. We then investigated the effect of endogenous NADPH availability on SA production.
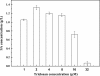
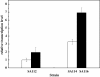
Results: The pps and csrB genes were first overexpressed by replacing their native promoter and integrating an additional copy of the genes in a double gene knockout (aroK and aroL) of E. coli. The aroG(fbr), aroB, aroE and tktA gene cluster was integrated into the above E. coli chromosome by direct transformation. The gene copy number was then evolved to the desired value by triclosan induction. The resulting strain, E. coli SA110, produced 8.9-fold more SA than did the parental strain E. coli (ΔaroKΔaroL). Following qRT-PCR analysis, another copy of the tktA gene under the control of the 5P(tac) promoter was inserted into the chromosome of E. coli SA110 to obtain the more productive strain E. coli SA110. Next, the NADPH availability was increased by overexpressing the pntAB or nadK genes, which further enhanced SA production. The final strain, E. coli SA116, produced 3.12 g/L of SA with a yield on glucose substrate of 0.33 mol/mol.
Conclusion: An SA-producing E. coli strain that carries neither a plasmid nor an antibiotic marker was constructed by triclosan-induced chromosomal evolution. We present the first demonstration that increasing NADPH availability by overexpressing the pntAB or nadK genes significantly enhances SA production.
Figures



Similar articles
-
Metabolic engineering for the production of shikimic acid in an evolved Escherichia coli strain lacking the phosphoenolpyruvate: carbohydrate phosphotransferase system.Microb Cell Fact. 2010 Apr 12;9:21. doi: 10.1186/1475-2859-9-21. Microb Cell Fact. 2010. PMID: 20385022 Free PMC article.
-
Improvement of shikimic acid production in Escherichia coli with growth phase-dependent regulation in the biosynthetic pathway from glycerol.World J Microbiol Biotechnol. 2017 Feb;33(2):25. doi: 10.1007/s11274-016-2192-3. Epub 2017 Jan 2. World J Microbiol Biotechnol. 2017. PMID: 28044275
-
Enhanced production of shikimic acid using a multi-gene co-expression system in Escherichia coli.Chin J Nat Med. 2016 Apr;14(4):286-293. doi: 10.1016/S1875-5364(16)30029-2. Chin J Nat Med. 2016. PMID: 27114316
-
[Improvements of shikimic acid production in Escherichia coli with ideal metabolic modification in biosynthetic pathway--a review].Wei Sheng Wu Xue Bao. 2014 Jan 4;54(1):5-13. Wei Sheng Wu Xue Bao. 2014. PMID: 24783849 Review. Chinese.
-
Metabolic engineering for microbial production of shikimic acid.Metab Eng. 2003 Oct;5(4):277-83. doi: 10.1016/j.ymben.2003.09.001. Metab Eng. 2003. PMID: 14642355 Review.
Cited by
-
Metabolic modeling and response surface analysis of an Escherichia coli strain engineered for shikimic acid production.BMC Syst Biol. 2018 Nov 12;12(1):102. doi: 10.1186/s12918-018-0632-4. BMC Syst Biol. 2018. PMID: 30419897 Free PMC article.
-
Advances and prospects in metabolic engineering of Escherichia coli for L-tryptophan production.World J Microbiol Biotechnol. 2022 Jan 6;38(2):22. doi: 10.1007/s11274-021-03212-1. World J Microbiol Biotechnol. 2022. PMID: 34989926 Review.
-
NAD Kinases: Metabolic Targets Controlling Redox Co-enzymes and Reducing Power Partitioning in Plant Stress and Development.Front Plant Sci. 2018 Mar 23;9:379. doi: 10.3389/fpls.2018.00379. eCollection 2018. Front Plant Sci. 2018. PMID: 29662499 Free PMC article. Review.
-
Metabolic engineering Saccharomyces cerevisiae for de novo biosynthesis of hydroxycinnamoyl glycerols.Synth Syst Biotechnol. 2025 Jul 17;10(4):1257-1266. doi: 10.1016/j.synbio.2025.07.005. eCollection 2025 Dec. Synth Syst Biotechnol. 2025. PMID: 40756073 Free PMC article.
-
Exploitation of Hetero- and Phototrophic Metabolic Modules for Redox-Intensive Whole-Cell Biocatalysis.Front Bioeng Biotechnol. 2022 Apr 13;10:855715. doi: 10.3389/fbioe.2022.855715. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 35497353 Free PMC article. Review.
References
-
- Kim CU, Lew W, Williams MA, Liu H, Zhang L, Swaminathan S, Bischofberger N, Chen MS, Mendel DB, Tai CY, Laver WG, Stevens RC. Influenza neuraminidase inhibitors possessing a novel hydrophobic interaction in the enzyme active site: design, synthesis, and structural analysis of carbocyclic sialic acid analogs with potent anti-influenza activity. J Am Chem Soc. 1997;119:681–690. doi: 10.1021/ja963036t. - DOI - PubMed
-
- Escalante A, Calderon R, Valdivia A, de Anda R, Hernández G, Hernández OT, Gosset G, Bolivar F. Metabolic engineering for the production of shikimic acid in an evolved Escherichia coli strain lacking the phosphoenolpyruvate: carbohydrate phosphotransferase system. Microb Cell Fact. 2010;9:21. doi: 10.1186/1475-2859-9-21. - DOI - PMC - PubMed
-
- Draths KM, Knop DR, Frost JW. Shikimic acid and quinic acid: replacing isolation from plant sources with recombinant microbial biocatalysis. J Am Chem Soc. 1999;121:1603–1604. doi: 10.1021/ja9830243. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

