A multi-split mapping algorithm for circular RNA, splicing, trans-splicing and fusion detection
- PMID: 24512684
- PMCID: PMC4056463
- DOI: 10.1186/gb-2014-15-2-r34
A multi-split mapping algorithm for circular RNA, splicing, trans-splicing and fusion detection
Abstract
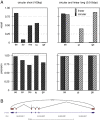
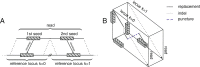
Numerous high-throughput sequencing studies have focused on detecting conventionally spliced mRNAs in RNA-seq data. However, non-standard RNAs arising through gene fusion, circularization or trans-splicing are often neglected. We introduce a novel, unbiased algorithm to detect splice junctions from single-end cDNA sequences. In contrast to other methods, our approach accommodates multi-junction structures. Our method compares favorably with competing tools for conventionally spliced mRNAs and, with a gain of up to 40% of recall, systematically outperforms them on reads with multiple splits, trans-splicing and circular products. The algorithm is integrated into our mapping tool segemehl (http://www.bioinf.uni-leipzig.de/Software/segemehl/).
Figures



References
-
- Frenkel-Morgenstern M, Lacroix V, Ezkurdia I, Levin Y, Gabashvili A, Prilusky J, del Pozo A, Tress M, Johnson R, Guigó R, Valencia A. Chimeras taking shape: potential functions of proteins encoded by chimeric RNA transcripts. Genome Res. 2012;22:1231–1242. doi: 10.1101/gr.130062.111. - DOI - PMC - PubMed
-
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- SRA/SRR018261
- SRA/SRR018262
- SRA/SRR166809
- SRA/SRX151602
LinkOut - more resources
Full Text Sources
Other Literature Sources

