Co-translational protein targeting to the bacterial membrane
- PMID: 24513458
- PMCID: PMC3999308
- DOI: 10.1016/j.bbamcr.2013.10.013
Co-translational protein targeting to the bacterial membrane
Abstract
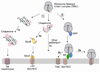
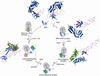
Co-translational protein targeting by the Signal Recognition Particle (SRP) is an essential cellular pathway that couples the synthesis of nascent proteins to their proper cellular localization. The bacterial SRP, which contains the minimal ribonucleoprotein core of this universally conserved targeting machine, has served as a paradigm for understanding the molecular basis of protein localization in all cells. In this review, we highlight recent biochemical and structural insights into the molecular mechanisms by which fundamental challenges faced by protein targeting machineries are met in the SRP pathway. Collectively, these studies elucidate how an essential SRP RNA and two regulatory GTPases in the SRP and SRP receptor (SR) enable this targeting machinery to recognize, sense and respond to its biological effectors, i.e. the cargo protein, the target membrane and the translocation machinery, thus driving efficient and faithful co-translational protein targeting. This article is part of a Special Issue entitled: Protein trafficking and secretion in bacteria. Guest Editors: Anastassios Economou and Ross Dalbey.
Keywords: GTPases; Molecular recognition and regulation; Protein targeting; Ribosome; SecYEG.
© 2013.
Figures
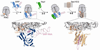

References
-
- Protein targeting, transport and translocation. London: Academic Press; 2002.
-
- Cross BCS, Sinning I, Luirink J, High S. Delivering proteins for export from the cytosol. Nat. Rev. Mol. Cell. Biol. 2009;10:255–264. - PubMed
-
- Hegde RS, Bernstein HD. The surprising complexity of signal sequences. Trends Biochem. Sci. 2006;31:563–571. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

