Coupling mRNA processing with transcription in time and space
- PMID: 24514444
- PMCID: PMC4304646
- DOI: 10.1038/nrg3662
Coupling mRNA processing with transcription in time and space
Abstract
Maturation of mRNA precursors often occurs simultaneously with their synthesis by RNA polymerase II (Pol II). The co-transcriptional nature of mRNA processing has permitted the evolution of coupling mechanisms that coordinate transcription with mRNA capping, splicing, editing and 3' end formation. Recent experiments using sophisticated new methods for analysis of nascent RNA have provided important insights into the relative amount of co-transcriptional and post-transcriptional processing, the relationship between mRNA elongation and processing, and the role of the Pol II carboxy-terminal domain (CTD) in regulating these processes.
Conflict of interest statement
Figures
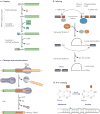
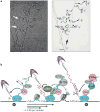
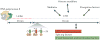
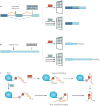
References
-
- Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. This is an early graphic demonstration of co-transcriptional splicing. - PubMed
-
- Bieberstein NI, Carrillo Oesterreich F, Straube K, Neugebauer KM. First exon length controls active chromatin signatures and transcription. Cell Rep. 2012;2:62–68. - PubMed
-
- Gomez Acuna LI, Fiszbein A, Allo M, Schor IE, Kornblihtt AR. Connections between chromatin signatures and splicing. Wiley Interdiscip Rev RNA. 2013;4:77–91. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

