Degradation of centromeric histone H3 variant Cse4 requires the Fpr3 peptidyl-prolyl Cis-Trans isomerase
- PMID: 24514906
- PMCID: PMC3982699
- DOI: 10.1534/genetics.114.161224
Degradation of centromeric histone H3 variant Cse4 requires the Fpr3 peptidyl-prolyl Cis-Trans isomerase
Abstract
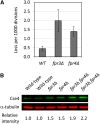
The centromeric histone H3 variant Cse4 in Saccharomyces cerevisiae is polyubiquitylated and degraded in a proteasome-dependent manner. We report here that the proline isomerase Fpr3 regulates Cse4 proteolysis. Structural change in Cse4 by Fpr3 might be important for the interaction between Cse4 and the E3 ubiquitin ligase Psh1.
Keywords: Cse4; Fpr3; Psh1; isomerization; protein degradation.
Figures



Similar articles
-
Molecular basis for the selective recognition and ubiquitination of centromeric histone H3 by yeast E3 ligase Psh1.J Genet Genomics. 2021 Jun 20;48(6):463-472. doi: 10.1016/j.jgg.2021.04.007. Epub 2021 May 26. J Genet Genomics. 2021. PMID: 34217622
-
Phosphorylation by casein kinase 2 facilitates Psh1 protein-assisted degradation of Cse4 protein.J Biol Chem. 2014 Oct 17;289(42):29297-309. doi: 10.1074/jbc.M114.580589. Epub 2014 Sep 2. J Biol Chem. 2014. PMID: 25183013 Free PMC article.
-
Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4.Mol Cell. 2010 Nov 12;40(3):444-54. doi: 10.1016/j.molcel.2010.10.014. Mol Cell. 2010. PMID: 21070970 Free PMC article.
-
Protein kinases in mitotic phosphorylation of budding yeast CENP-A.Curr Genet. 2019 Dec;65(6):1325-1332. doi: 10.1007/s00294-019-00997-5. Epub 2019 May 22. Curr Genet. 2019. PMID: 31119371 Review.
-
Dynamic ubiquitin signaling in cell cycle regulation.J Cell Biol. 2017 Aug 7;216(8):2259-2271. doi: 10.1083/jcb.201703170. Epub 2017 Jul 6. J Cell Biol. 2017. PMID: 28684425 Free PMC article. Review.
Cited by
-
Recent insights into mechanisms preventing ectopic centromere formation.Open Biol. 2021 Sep;11(9):210189. doi: 10.1098/rsob.210189. Epub 2021 Sep 8. Open Biol. 2021. PMID: 34493071 Free PMC article. Review.
-
Reduced gene dosage of histone H4 prevents CENP-A mislocalization and chromosomal instability in Saccharomyces cerevisiae.Genetics. 2021 May 17;218(1):iyab033. doi: 10.1093/genetics/iyab033. Genetics. 2021. PMID: 33751052 Free PMC article.
-
Conformational flexibility of histone variant CENP-ACse4 is regulated by histone H4: A mechanism to stabilize soluble Cse4.J Biol Chem. 2018 Dec 28;293(52):20273-20284. doi: 10.1074/jbc.RA118.004141. Epub 2018 Oct 31. J Biol Chem. 2018. PMID: 30381395 Free PMC article.
-
Cdc7-mediated phosphorylation of Cse4 regulates high-fidelity chromosome segregation in budding yeast.Mol Biol Cell. 2021 Nov 1;32(21):ar15. doi: 10.1091/mbc.E21-06-0323. Epub 2021 Aug 25. Mol Biol Cell. 2021. PMID: 34432494 Free PMC article.
-
CENP-A overexpression promotes aneuploidy with karyotypic heterogeneity.J Cell Biol. 2021 Apr 5;220(4):e202007195. doi: 10.1083/jcb.202007195. J Cell Biol. 2021. PMID: 33620383 Free PMC article.
References
-
- Arevalo-Rodriguez M., Wu X., Hanes S. D., Heitman J., 2004. Prolyl isomerases in yeast. Front. Biosci. 9: 2420–2446. - PubMed
-
- Collins S. R., Miller K. M., Maas N. L., Roguev A., Fillingham J., et al. , 2007. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446: 806–810. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

