T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow
- PMID: 24516120
- PMCID: PMC3949572
- DOI: 10.1084/jem.20131560
T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow
Abstract
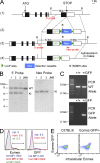
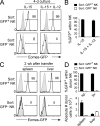
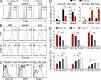
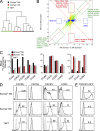
Trail(+)DX5(-)Eomes(-) natural killer (NK) cells arise in the mouse fetal liver and persist in the adult liver. Their relationships with Trail(-)DX5(+) NK cells remain controversial. We generated a novel Eomes-GFP reporter murine model to address this question. We found that Eomes(-) NK cells are not precursors of classical Eomes(+) NK cells but rather constitute a distinct lineage of innate lymphoid cells. Eomes(-) NK cells are strictly dependent on both T-bet and IL-15, similarly to NKT cells. We observed that, in the liver, expression of T-bet in progenitors represses Eomes expression and the development of Eomes(+) NK cells. Reciprocally, the bone marrow (BM) microenvironment restricts T-bet expression in developing NK cells. Ectopic expression of T-bet forces the development of Eomes(-) NK cells, demonstrating that repression of T-bet is essential for the development of Eomes(+) NK cells. Gene profile analyses show that Eomes(-) NK cells share part of their transcriptional program with NKT cells, including genes involved in liver homing and NK cell receptors. Moreover, Eomes(-) NK cells produce a broad range of cytokines, including IL-2 and TNF in vitro and in vivo, during immune responses against vaccinia virus. Thus, mutually exclusive expression of T-bet and Eomes drives the development of different NK cell lineages with complementary functions.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

