NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly
- PMID: 24516400
- PMCID: PMC3916286
- DOI: 10.1371/journal.pgen.1004110
NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly
Abstract
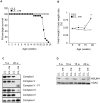
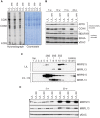
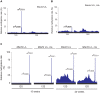
Biogenesis of mammalian mitochondrial ribosomes requires a concerted maturation of both the small (SSU) and large subunit (LSU). We demonstrate here that the m(5)C methyltransferase NSUN4, which forms a complex with MTERF4, is essential in mitochondrial ribosomal biogenesis as mitochondrial translation is abolished in conditional Nsun4 mouse knockouts. Deep sequencing of bisulfite-treated RNA shows that NSUN4 methylates cytosine 911 in 12S rRNA (m5C911) of the SSU. Surprisingly, NSUN4 does not need MTERF4 to generate this modification. Instead, the NSUN4/MTERF4 complex is required to assemble the SSU and LSU to form a monosome. NSUN4 is thus a dual function protein, which on the one hand is needed for 12S rRNA methylation and, on the other hand interacts with MTERF4 to facilitate monosome assembly. The presented data suggest that NSUN4 has a key role in controlling a final step in ribosome biogenesis to ensure that only the mature SSU and LSU are assembled.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Structure of the human MTERF4-NSUN4 protein complex that regulates mitochondrial ribosome biogenesis.Proc Natl Acad Sci U S A. 2012 Sep 18;109(38):15253-8. doi: 10.1073/pnas.1210688109. Epub 2012 Sep 4. Proc Natl Acad Sci U S A. 2012. PMID: 22949673 Free PMC article.
-
MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome.Cell Metab. 2011 May 4;13(5):527-39. doi: 10.1016/j.cmet.2011.04.002. Cell Metab. 2011. PMID: 21531335
-
Structure of the essential MTERF4:NSUN4 protein complex reveals how an MTERF protein collaborates to facilitate rRNA modification.Structure. 2012 Nov 7;20(11):1940-7. doi: 10.1016/j.str.2012.08.027. Epub 2012 Sep 27. Structure. 2012. PMID: 23022348 Free PMC article.
-
Coupling of import and assembly pathways in mitochondrial protein biogenesis.Biol Chem. 2019 Dec 18;401(1):117-129. doi: 10.1515/hsz-2019-0310. Biol Chem. 2019. PMID: 31513529 Review.
-
Epitranscriptomics of Mammalian Mitochondrial Ribosomal RNA.Cells. 2020 Sep 27;9(10):2181. doi: 10.3390/cells9102181. Cells. 2020. PMID: 32992603 Free PMC article. Review.
Cited by
-
Human Mitoribosome Biogenesis and Its Emerging Links to Disease.Int J Mol Sci. 2021 Apr 7;22(8):3827. doi: 10.3390/ijms22083827. Int J Mol Sci. 2021. PMID: 33917098 Free PMC article. Review.
-
Testing a Hypothesis of 12S rRNA Methylation by Putative METTL17 Methyltransferase.Acta Naturae. 2023 Oct-Dec;15(4):75-82. doi: 10.32607/actanaturae.25441. Acta Naturae. 2023. PMID: 38234605 Free PMC article.
-
The potential of RNA methylation in the treatment of cardiovascular diseases.iScience. 2024 Jul 20;27(8):110524. doi: 10.1016/j.isci.2024.110524. eCollection 2024 Aug 16. iScience. 2024. PMID: 39165846 Free PMC article. Review.
-
Mitoribosome structure with cofactors and modifications reveals mechanism of ligand binding and interactions with L1 stalk.Nat Commun. 2024 May 20;15(1):4272. doi: 10.1038/s41467-024-48163-x. Nat Commun. 2024. PMID: 38769321 Free PMC article.
-
Human Mitochondrial RNA Processing and Modifications: Overview.Int J Mol Sci. 2021 Jul 27;22(15):7999. doi: 10.3390/ijms22157999. Int J Mol Sci. 2021. PMID: 34360765 Free PMC article. Review.
References
-
- Cavdar Koc E, Burkhart W, Blackburn K, Moseley A, Spremulli LL (2001) The small subunit of the mammalian mitochondrial ribosome. Identification of the full complement of ribosomal proteins present. J Biol Chem 276: 19363–19374. - PubMed
-
- Koc EC, Burkhart W, Blackburn K, Moyer MB, Schlatzer DM, et al. (2001) The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J Biol Chem 276: 43958–43969. - PubMed
-
- Metodiev MD, Lesko N, Park CB, Camara Y, Shi Y, et al. (2009) Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab 9: 386–397. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

