Mechanistically distinct mouse models for CRX-associated retinopathy
- PMID: 24516401
- PMCID: PMC3916252
- DOI: 10.1371/journal.pgen.1004111
Mechanistically distinct mouse models for CRX-associated retinopathy
Abstract
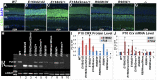
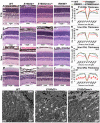
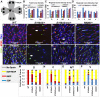
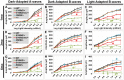
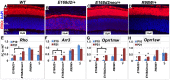
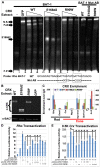
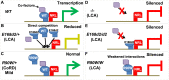
Cone-rod homeobox (CRX) protein is a "paired-like" homeodomain transcription factor that is essential for regulating rod and cone photoreceptor transcription. Mutations in human CRX are associated with the dominant retinopathies Retinitis Pigmentosa (RP), Cone-Rod Dystrophy (CoRD) and Leber Congenital Amaurosis (LCA), with variable severity. Heterozygous Crx Knock-Out (KO) mice ("+/-") have normal vision as adults and fail to model the dominant human disease. To investigate how different mutant CRX proteins produce distinct disease pathologies, we generated two Crx Knock-IN (K-IN) mouse models: Crx(E168d2) ("E168d2") and Crx(R90W) ("R90W"). E168d2 mice carry a frameshift mutation in the CRX activation domain, Glu168del2, which is associated with severe dominant CoRD or LCA in humans. R90W mice carry a substitution mutation in the CRX homeodomain, Arg90Trp, which is associated with dominant mild late-onset CoRD and recessive LCA. As seen in human patients, heterozygous E168d2 ("E168d2/+") but not R90W ("R90W/+") mice show severely impaired retinal function, while mice homozygous for either mutation are blind and undergo rapid photoreceptor degeneration. E168d2/+ mice also display abnormal rod/cone morphology, greater impairment of CRX target gene expression than R90W/+ or +/- mice, and undergo progressive photoreceptor degeneration. Surprisingly, E168d2/+ mice express more mutant CRX protein than wild-type CRX. E168d2neo/+, a subline of E168d2 with reduced mutant allele expression, displays a much milder retinal phenotype, demonstrating the impact of Crx expression level on disease severity. Both CRX([E168d2]) and CRX([R90W]) proteins fail to activate transcription in vitro, but CRX([E168d2]) interferes more strongly with the function of wild type (WT) CRX, supporting an antimorphic mechanism. E168d2 and R90W are mechanistically distinct mouse models for CRX-associated disease that will allow the elucidation of molecular mechanisms and testing of novel therapeutic approaches for different forms of CRX-associated disease.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Chen S, Wang QL, Nie Z, Sun H, Lennon G, et al. (1997) Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron 19(5): 1017–30. - PubMed
-
- Furukawa T, Morrow EM, Cepko CL (1997) Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell 91: 531–541. - PubMed
-
- Furukawa T, Morrow EM, Li T, Davis FC, Cepko CL (1999) Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat Genet 23 4: 466–470. - PubMed
-
- Chau KY, Chen S, Zack DJ, Ono SJ (2000) Functional domains of the cone-rod homeobox (CRX) transcription factor. The Journal of biological chemistry 275 47: 37264–70. - PubMed
-
- Kimura A, Singh D, Wawrousek E, Kikuchi M, Nakamura M, et al. (2000) Both PCE-1/RX and OTX/CRX Interactions Are Necessary for Photoreceptor-specific Gene Expression. J Biol Chem 275: 1152–1160. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

