Metabolic evolution of a deep-branching hyperthermophilic chemoautotrophic bacterium
- PMID: 24516572
- PMCID: PMC3917532
- DOI: 10.1371/journal.pone.0087950
Metabolic evolution of a deep-branching hyperthermophilic chemoautotrophic bacterium
Erratum in
- PLoS One. 2014;9(3):e93345
Abstract
Aquifex aeolicus is a deep-branching hyperthermophilic chemoautotrophic bacterium restricted to hydrothermal vents and hot springs. These characteristics make it an excellent model system for studying the early evolution of metabolism. Here we present the whole-genome metabolic network of this organism and examine in detail the driving forces that have shaped it. We make extensive use of phylometabolic analysis, a method we recently introduced that generates trees of metabolic phenotypes by integrating phylogenetic and metabolic constraints. We reconstruct the evolution of a range of metabolic sub-systems, including the reductive citric acid (rTCA) cycle, as well as the biosynthesis and functional roles of several amino acids and cofactors. We show that A. aeolicus uses the reconstructed ancestral pathways within many of these sub-systems, and highlight how the evolutionary interconnections between sub-systems facilitated several key innovations. Our analyses further highlight three general classes of driving forces in metabolic evolution. One is the duplication and divergence of genes for enzymes as these progress from lower to higher substrate specificity, improving the kinetics of certain sub-systems. A second is the kinetic optimization of established pathways through fusion of enzymes, or their organization into larger complexes. The third is the minimization of the ATP unit cost to synthesize biomass, improving thermodynamic efficiency. Quantifying the distribution of these classes of innovations across metabolic sub-systems and across the tree of life will allow us to assess how a tradeoff between maximizing growth rate and growth efficiency has shaped the long-term metabolic evolution of the biosphere.
Conflict of interest statement
Figures
 or
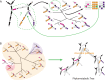
or  position of THF remains to be elucidated (see text). Relative to the reconstructed root of carbon-fixation, in which the rTCA cycle and Wood-Ljungdahl pathway are fully integrated , this hybrid strategy employed by A. aeolicus lacks only the grey-dashed reaction (acetyl-CoA synthesis). Molecules highlighted in blue represent the “pillars of anabolism”, TCA intermediates from which the vast majority of anabolic pathways have been initiated throughout evolution . Highlighted in green is succinyl-CoA, which forms a precursor to pyrroles through a later derived pathway in some organisms (but not A. aeolicus). Highlighted in red are reaction sequences involving the same local functional group transformation that in A. aeolicus are catalyzed by closely related enzymes in both halves of the rTCA cycle. Green dashed arrows highlight alternate pathway sequences catalyzed by a single enzyme in other clades.
position of THF remains to be elucidated (see text). Relative to the reconstructed root of carbon-fixation, in which the rTCA cycle and Wood-Ljungdahl pathway are fully integrated , this hybrid strategy employed by A. aeolicus lacks only the grey-dashed reaction (acetyl-CoA synthesis). Molecules highlighted in blue represent the “pillars of anabolism”, TCA intermediates from which the vast majority of anabolic pathways have been initiated throughout evolution . Highlighted in green is succinyl-CoA, which forms a precursor to pyrroles through a later derived pathway in some organisms (but not A. aeolicus). Highlighted in red are reaction sequences involving the same local functional group transformation that in A. aeolicus are catalyzed by closely related enzymes in both halves of the rTCA cycle. Green dashed arrows highlight alternate pathway sequences catalyzed by a single enzyme in other clades.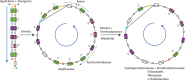
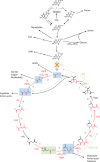
 , while fusion of one sub-unit of enzyme 1 with enzyme for the subsequent cleavage reaction improves kinetics. Green boxes represent homologous reactions catalyzed by enzymes with high sequence similarity, while purple boxes represent homologous reactions catalyzed by members of the same enzyme families. Reactions 5′ and 5* are catalyzed by the same enzyme. Differences in sequence divergence between green and purple enzymes may reflect differences in complexity of the reactions, see text for further discussion. The yellow node represents acetyl-CoA, the blue node represents oxaloacetate, and the red node represents succinyl-CoA. Dark blue arrows indicate the direction of mass through pathways.
, while fusion of one sub-unit of enzyme 1 with enzyme for the subsequent cleavage reaction improves kinetics. Green boxes represent homologous reactions catalyzed by enzymes with high sequence similarity, while purple boxes represent homologous reactions catalyzed by members of the same enzyme families. Reactions 5′ and 5* are catalyzed by the same enzyme. Differences in sequence divergence between green and purple enzymes may reflect differences in complexity of the reactions, see text for further discussion. The yellow node represents acetyl-CoA, the blue node represents oxaloacetate, and the red node represents succinyl-CoA. Dark blue arrows indicate the direction of mass through pathways.
 -ketobutyrate synthesis, reconstructed here to represent the ancestral sequence to this compound. The molecules highlighted in orange and green in turn show the compact interconnectedness of the ancestral pathways to the branched chain amino acids. Parallels to substrate sequences within the oxidative TCA are also highlighted, as well as the alternate route to
-ketobutyrate synthesis, reconstructed here to represent the ancestral sequence to this compound. The molecules highlighted in orange and green in turn show the compact interconnectedness of the ancestral pathways to the branched chain amino acids. Parallels to substrate sequences within the oxidative TCA are also highlighted, as well as the alternate route to  -ketobutyrate from threonine.
-ketobutyrate from threonine.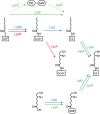
References
-
- Braakman R, Smith E (2013) The compositional and evolutionary logic of metabolism. Physical Biology 10: 011001. - PubMed
-
- Huber R, Eder W (2006) Aquificales. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editors, The Prokaryotes, Springer New York. 925–938.
-
- Reysenbach AL, Shock E (2002) Merging genomes with geochemistry in hydrothermal ecosystems. Science 296: 1077–1082. - PubMed
-
- Guiral M, Prunetti L, Aussignargues C, Ciaccafava A, Infossi P, et al. (2012) The hyperthermophilic bacterium Aquifex aeolicus: From respiratory pathways to extremely resistant enzymes and biotechnological applications. Advances in Microbial Physiology 61: 125. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

