Behavioral signatures related to genetic disorders in autism
- PMID: 24517317
- PMCID: PMC3936826
- DOI: 10.1186/2040-2392-5-11
Behavioral signatures related to genetic disorders in autism
Abstract
Background: Autism spectrum disorder (ASD) is well recognized to be genetically heterogeneous. It is assumed that the genetic risk factors give rise to a broad spectrum of indistinguishable behavioral presentations.
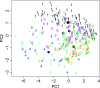
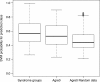
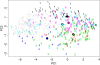
Methods: We tested this assumption by analyzing the Autism Diagnostic Interview-Revised (ADI-R) symptom profiles in samples comprising six genetic disorders that carry an increased risk for ASD (22q11.2 deletion, Down's syndrome, Prader-Willi, supernumerary marker chromosome 15, tuberous sclerosis complex and Klinefelter syndrome; total n = 322 cases, groups ranging in sample sizes from 21 to 90 cases). We mined the data to test the existence and specificity of ADI-R profiles using a multiclass extension of support vector machine (SVM) learning. We subsequently applied the SVM genetic disorder algorithm on idiopathic ASD profiles from the Autism Genetics Resource Exchange (AGRE).
Results: Genetic disorders were associated with behavioral specificity, indicated by the accuracy and certainty of SVM predictions; one-by-one genetic disorder stratifications were highly accurate leading to 63% accuracy of correct genotype prediction when all six genetic disorder groups were analyzed simultaneously. Application of the SVM algorithm to AGRE cases indicated that the algorithm could detect similarity of genetic behavioral signatures in idiopathic ASD subjects. Also, affected sib pairs in the AGRE were behaviorally more similar when they had been allocated to the same genetic disorder group.
Conclusions: Our findings provide evidence for genotype-phenotype correlations in relation to autistic symptomatology. SVM algorithms may be used to stratify idiopathic cases of ASD according to behavioral signature patterns associated with genetic disorders. Together, the results suggest a new approach for disentangling the heterogeneity of ASD.
Figures

References
-
- Betancur C. Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain Res. 2011;1380:42–77. - PubMed
-
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, Almeida J, Bacchelli E, Bader GD, Bailey AJ, Baird G, Battaglia A, Berney T, Bolshakova N, Bölte S, Bolton PF, Bourgeron T, Brennan S, Brian J, Bryson SE, Carson AR, Casallo G, Casey J, Chung BH, Cochrane L, Corsello C. et al.Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466(7304):368–372. doi: 10.1038/nature09146. - DOI - PMC - PubMed
-
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, Chu SH, Moreau MP, Gupta AR, Thomson SA, Mason CE, Bilguvar K, Celestino-Soper PB, Choi M, Crawford EL, Davis L, Wright NR, Dhodapkar RM, DiCola M, DiLullo NM, Fernandez TV, Fielding-Singh V, Fishman DO, Frahm S, Garagaloyan R, Goh GS, Kammela S, Klei L, Lowe JK, Lund SC. et al.Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70(5):863–885. doi: 10.1016/j.neuron.2011.05.002. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

