Discovery of new membrane-associated proteins overexpressed in small-cell lung cancer
- PMID: 24518087
- PMCID: PMC4028683
- DOI: 10.1097/JTO.0000000000000090
Discovery of new membrane-associated proteins overexpressed in small-cell lung cancer
Abstract
Introduction: Small-cell lung cancer (SCLC) is the most aggressive subtype of lung cancer, with no early detection strategy or targeted therapy currently available. We hypothesized that difference gel electrophoresis (DIGE) may identify membrane-associated proteins (MAPs) specific to SCLC, advance our understanding of SCLC biology, and discover new biomarkers of SCLC.
Methods: MAP lysates were prepared from three SCLCs, three non-small-cell lung cancers, and three immortalized normal bronchial epithelial cell lines and coanalyzed by DIGE. Subsequent protein identification was performed by mass spectrometry. Proteins were submitted to Ingenuity Pathway Analysis. Candidate biomarkers were validated by Western blotting (WB) and immunohistochemistry (IHC).
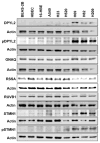
Results: Principal component analysis on the global DIGE data set demonstrated that the four replicates derived from each of the nine cell lines clustered closely, as did samples within the same histological group. One hundred thirty-seven proteins were differentially expressed in SCLC compared with non-small-cell lung cancer and immortalized normal bronchial epithelial cells. These proteins were overrepresented in cellular/tissue morphology networks. Dihydropyrimidinase-related protein 2, guanine nucleotide-binding protein alpha-q, laminin receptor 1, pontin, and stathmin 1 were selected as candidate biomarkers among MAPs overexpressed in SCLC. Overexpression of all candidates but RSSA in SCLC was verified by WB and/or IHC on tissue microarrays. These proteins were significantly associated with SCLC histology and survival in univariables analyses.
Conclusion: DIGE analysis of a membrane-associated subproteome discovered overexpression of dihydropyrimidinase-related protein 2, guanine nucleotide-binding protein alpha-q, RUVB1, and stathmin 1 in SCLC. Results were verified by WB and/or IHC in primary tumors, suggesting that investigating their functional relevance in SCLC progression is warranted. Association with survival requires further validation in larger clinical data sets.
Conflict of interest statement
Disclosure: The authors declare no conflict of interest.
Figures



Similar articles
-
Identification of isocitrate dehydrogenase 1 as a potential diagnostic and prognostic biomarker for non-small cell lung cancer by proteomic analysis.Mol Cell Proteomics. 2012 Feb;11(2):M111.008821. doi: 10.1074/mcp.M111.008821. Epub 2011 Nov 7. Mol Cell Proteomics. 2012. PMID: 22064513 Free PMC article.
-
Acyl-coenzyme A-binding protein regulates Beta-oxidation required for growth and survival of non-small cell lung cancer.Cancer Prev Res (Phila). 2014 Jul;7(7):748-57. doi: 10.1158/1940-6207.CAPR-14-0057. Epub 2014 May 12. Cancer Prev Res (Phila). 2014. PMID: 24819876 Free PMC article.
-
An immunohistochemical study of cyclin-dependent kinase 5 (CDK5) expression in non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC): a possible prognostic biomarker.World J Surg Oncol. 2016 Feb 9;14(1):34. doi: 10.1186/s12957-016-0787-7. World J Surg Oncol. 2016. PMID: 26860827 Free PMC article.
-
Update on small cell carcinoma and its differentiation from squamous cell carcinoma and other non-small cell carcinomas.Mod Pathol. 2012 Jan;25 Suppl 1:S18-30. doi: 10.1038/modpathol.2011.150. Mod Pathol. 2012. PMID: 22214967 Review.
-
An overview of mortality & predictors of small-cell and non-small cell lung cancer among Saudi patients.J Epidemiol Glob Health. 2018 Mar;7 Suppl 1(Suppl 1):S1-S6. doi: 10.1016/j.jegh.2017.09.004. Epub 2017 Sep 28. J Epidemiol Glob Health. 2018. PMID: 29801587 Free PMC article. Review.
Cited by
-
Telomerase and Telomeres in Endometrial Cancer.Front Oncol. 2019 May 17;9:344. doi: 10.3389/fonc.2019.00344. eCollection 2019. Front Oncol. 2019. PMID: 31157162 Free PMC article. Review.
-
The Role of Pontin and Reptin in Cellular Physiology and Cancer Etiology.Front Mol Biosci. 2017 Aug 24;4:58. doi: 10.3389/fmolb.2017.00058. eCollection 2017. Front Mol Biosci. 2017. PMID: 28884116 Free PMC article. Review.
-
Lung cancer research is taking on new challenges: knowledge of tumors' molecular diversity is opening new pathways to treatment.P T. 2014 Oct;39(10):698-714. P T. 2014. PMID: 25336866 Free PMC article.
-
Downregulation of RUVBL1 inhibits proliferation of lung adenocarcinoma cells by G1/S phase cell cycle arrest via multiple mechanisms.Tumour Biol. 2016 Dec;37:16015–16027. doi: 10.1007/s13277-016-5452-9. Epub 2016 Oct 10. Tumour Biol. 2016. PMID: 27722820
-
"Stealth dissemination" of macrophage-tumor cell fusions cultured from blood of patients with pancreatic ductal adenocarcinoma.PLoS One. 2017 Sep 28;12(9):e0184451. doi: 10.1371/journal.pone.0184451. eCollection 2017. PLoS One. 2017. PMID: 28957348 Free PMC article.
References
-
- Jackman DM, Johnson BE. Small-cell lung cancer. Lancet. 2005;366:1385–1396. - PubMed
-
- Alfonso P, Núñez A, Madoz-Gurpide J, Lombardia L, Sánchez L, Casal JI. Proteomic expression analysis of colorectal cancer by two-dimensional differential gel electrophoresis. Proteomics. 2005;5:2602–2611. - PubMed
-
- Evans CA, Tonge R, Blinco D, et al. Comparative proteomics of primitive hematopoietic cell populations reveals differences in expression of proteins regulating motility. Blood. 2004;103:3751–3759. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

