Identification of a long non-coding RNA as a novel biomarker and potential therapeutic target for metastatic prostate cancer
- PMID: 24519926
- PMCID: PMC3996663
- DOI: 10.18632/oncotarget.1769
Identification of a long non-coding RNA as a novel biomarker and potential therapeutic target for metastatic prostate cancer
Abstract
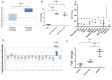
Metastatic prostate cancer (PCa) is still an incurable disease. Long non-coding RNAs (lncRNAs) may be an overlooked source of cancer biomarkers and therapeutic targets. We therefore performed RNA sequencing on paired metastatic/non-metastatic PCa xenografts derived from clinical specimens. The most highly up-regulated transcript was LOC728606, a lncRNA now designated PCAT18. PCAT18 is specifically expressed in the prostate compared to 11 other normal tissues (p<0.05) and up-regulated in PCa compared to 15 other neoplasms (p<0.001). Cancer-specific up-regulation of PCAT18 was confirmed on an independent dataset of PCa and benign prostatic hyperplasia samples (p<0.001). PCAT18 was detectable in plasma samples and increased incrementally from healthy individuals to those with localized and metastatic PCa (p<0.01). We identified a PCAT18-associated expression signature (PES), which is highly PCa-specific and activated in metastatic vs. primary PCa samples (p<1E-4, odds ratio>2). The PES was significantly associated with androgen receptor (AR) signalling. Accordingly, AR activation dramatically up-regulated PCAT18 expression in vitro and in vivo. PCAT18 silencing significantly (p<0.001) inhibited PCa cell proliferation and triggered caspase 3/7 activation, with no effect on non-neoplastic cells. PCAT18 silencing also inhibited PCa cell migration (p<0.01) and invasion (p<0.01). These results position PCAT18 as a potential therapeutic target and biomarker for metastatic PCa.
Conflict of interest statement
No potential conflicts of interest were declared.
Figures



References
-
- Bishr M, Saad F. Overview of the latest treatments for castration-resistant prostate cancer. Nat Rev Urol. 2013 - PubMed
-
- Rodrigues G, Warde P, Pickles T, Crook J, Brundage M, Souhami L, Lukka H, Genitourinary Radiation Oncologists of C Pre-treatment risk stratification of prostate cancer patients: A critical review. Canadian Urological Association journal = Journal de l'Association des urologues du Canada. 2012;6(2):121–127. - PMC - PubMed
-
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484–1488. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

