The SMC complex MukBEF recruits topoisomerase IV to the origin of replication region in live Escherichia coli
- PMID: 24520061
- PMCID: PMC3950513
- DOI: 10.1128/mBio.01001-13
The SMC complex MukBEF recruits topoisomerase IV to the origin of replication region in live Escherichia coli
Abstract
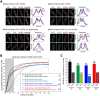
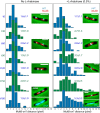
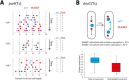
The Escherichia coli structural maintenance of chromosome (SMC) complex, MukBEF, and topoisomerase IV (TopoIV) interact in vitro through a direct contact between the MukB dimerization hinge and the C-terminal domain of ParC, the catalytic subunit of TopoIV. The interaction stimulates catalysis by TopoIV in vitro. Using live-cell quantitative imaging, we show that MukBEF directs TopoIV to ori, with fluorescent fusions of ParC and ParE both forming cellular foci that colocalize with those formed by MukBEF throughout the cell cycle and in cells unable to initiate DNA replication. Removal of MukBEF leads to loss of fluorescent ParC/ParE foci. In the absence of functional TopoIV, MukBEF forms multiple foci that are distributed uniformly throughout the nucleoid, whereas multiple catenated oris cluster at midcell. Once functional TopoIV is restored, the decatenated oris segregate to positions that are largely coincident with the MukBEF foci, thereby providing support for a mechanism by which MukBEF acts in chromosome segregation by positioning newly replicated and decatenated oris. Additional evidence for such a mechanism comes from the observation that in TopoIV-positive (TopoIV(+)) cells, newly replicated oris segregate rapidly to the positions of MukBEF foci. Taken together, the data implicate MukBEF as a key component of the DNA segregation process by acting in concert with TopoIV to promote decatenation and positioning of newly replicated oris.
Importance: Mechanistic understanding of how newly replicated bacterial chromosomes are segregated prior to cell division is incomplete. In this work, we provide in vivo experimental support for the view that topoisomerase IV (TopoIV), which decatenates newly replicated sister duplexes as a prelude to successful segregation, is directed to the replication origin region of the Escherichia coli chromosome by the SMC (structural maintenance of chromosome) complex, MukBEF. We provide in vivo data that support the demonstration in vitro that the MukB interaction with TopoIV stimulates catalysis by TopoIV. Finally, we show that MukBEF directs the normal positioning of sister origins after their replication and during their segregation. Overall, the data support models in which the coordinate and sequential action of TopoIV and MukBEF plays an important role during bacterial chromosome segregation.
Figures

Similar articles
-
MukBEF, a chromosomal organizer.J Mol Microbiol Biotechnol. 2014;24(5-6):371-83. doi: 10.1159/000369099. Epub 2015 Feb 17. J Mol Microbiol Biotechnol. 2014. PMID: 25732339 Free PMC article. Review.
-
The Localization and Action of Topoisomerase IV in Escherichia coli Chromosome Segregation Is Coordinated by the SMC Complex, MukBEF.Cell Rep. 2015 Dec 22;13(11):2587-2596. doi: 10.1016/j.celrep.2015.11.034. Epub 2015 Dec 10. Cell Rep. 2015. PMID: 26686641 Free PMC article.
-
MatP regulates the coordinated action of topoisomerase IV and MukBEF in chromosome segregation.Nat Commun. 2016 Jan 28;7:10466. doi: 10.1038/ncomms10466. Nat Commun. 2016. PMID: 26818444 Free PMC article.
-
The Escherichia coli SMC complex, MukBEF, shapes nucleoid organization independently of DNA replication.J Bacteriol. 2012 Sep;194(17):4669-76. doi: 10.1128/JB.00957-12. Epub 2012 Jun 29. J Bacteriol. 2012. PMID: 22753058 Free PMC article.
-
SMC complexes organize the bacterial chromosome by lengthwise compaction.Curr Genet. 2020 Oct;66(5):895-899. doi: 10.1007/s00294-020-01076-w. Epub 2020 Apr 16. Curr Genet. 2020. PMID: 32300862 Free PMC article. Review.
Cited by
-
MukB ATPases are regulated independently by the N- and C-terminal domains of MukF kleisin.Elife. 2018 Jan 11;7:e31522. doi: 10.7554/eLife.31522. Elife. 2018. PMID: 29323635 Free PMC article.
-
What makes a type IIA topoisomerase a gyrase or a Topo IV?Nucleic Acids Res. 2021 Jun 21;49(11):6027-6042. doi: 10.1093/nar/gkab270. Nucleic Acids Res. 2021. PMID: 33905522 Free PMC article. Review.
-
MukBEF, a chromosomal organizer.J Mol Microbiol Biotechnol. 2014;24(5-6):371-83. doi: 10.1159/000369099. Epub 2015 Feb 17. J Mol Microbiol Biotechnol. 2014. PMID: 25732339 Free PMC article. Review.
-
Chromosome Segregation in Bacillus subtilis Follows an Overall Pattern of Linear Movement and Is Highly Robust against Cell Cycle Perturbations.mSphere. 2020 Jun 17;5(3):e00255-20. doi: 10.1128/mSphere.00255-20. mSphere. 2020. PMID: 32554717 Free PMC article.
-
Mid-cell migration of the chromosomal terminus is coupled to origin segregation in Escherichia coli.Nat Commun. 2023 Nov 18;14(1):7489. doi: 10.1038/s41467-023-43351-7. Nat Commun. 2023. PMID: 37980336 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
