Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers
- PMID: 24520176
- PMCID: PMC3939885
- DOI: 10.1073/pnas.1316793111
Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers
Abstract
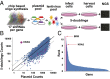
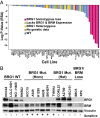
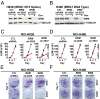
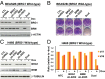
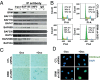
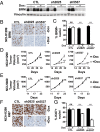
Defects in epigenetic regulation play a fundamental role in the development of cancer, and epigenetic regulators have recently emerged as promising therapeutic candidates. We therefore set out to systematically interrogate epigenetic cancer dependencies by screening an epigenome-focused deep-coverage design shRNA (DECODER) library across 58 cancer cell lines. This screen identified BRM/SMARCA2, a DNA-dependent ATPase of the mammalian SWI/SNF (mSWI/SNF) chromatin remodeling complex, as being essential for the growth of tumor cells that harbor loss of function mutations in BRG1/SMARCA4. Depletion of BRM in BRG1-deficient cancer cells leads to a cell cycle arrest, induction of senescence, and increased levels of global H3K9me3. We further demonstrate the selective dependency of BRG1-mutant tumors on BRM in vivo. Genetic alterations of the mSWI/SNF chromatin remodeling complexes are the most frequent among chromatin regulators in cancers, with BRG1/SMARCA4 mutations occurring in ∼10-15% of lung adenocarcinomas. Our findings position BRM as an attractive therapeutic target for BRG1 mutated cancers. Because BRG1 and BRM function as mutually exclusive catalytic subunits of the mSWI/SNF complex, we propose that such synthetic lethality may be explained by paralog insufficiency, in which loss of one family member unveils critical dependence on paralogous subunits. This concept of "cancer-selective paralog dependency" may provide a more general strategy for targeting other tumor suppressor lesions/complexes with paralogous subunits.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11(7):481–492. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

