Galactoside-binding site in LacY
- PMID: 24520888
- PMCID: PMC3985706
- DOI: 10.1021/bi401716z
Galactoside-binding site in LacY
Abstract
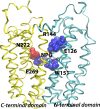
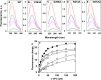
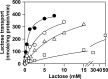
Although an X-ray crystal structure of lactose permease (LacY) has been presented with bound galactopyranoside, neither the sugar nor the residues ligating the sugar can be identified with precision at ~3.5 Å. Therefore, additional evidence is important for identifying side chains likely to be involved in binding. On the basis of a clue from site-directed alkylation suggesting that Asn272, Gly268, and Val264 on one face of helix VIII might participate in galactoside binding, molecular dynamics simulations were conducted initially. The simulations indicate that Asn272 (helix VIII) is sufficiently close to the galactopyranosyl ring of a docked lactose analogue to play an important role in binding, the backbone at Gly268 may be involved, and Val264 does not interact with the bound sugar. When the three side chains are subjected to site-directed mutagenesis, with the sole exception of mutant Asn272 → Gln, various other replacements for Asn272 either markedly decrease affinity for the substrate (i.e., high KD) or abolish binding altogether. However, mutant Gly268 → Ala exhibits a moderate 8-fold decrease in affinity, and binding by mutant Val264 → Ala is affected only minimally. Thus, Asn272 and possibly Gly268 may comprise additional components of the galactoside-binding site in LacY.
Figures


References
-
- Madej M. G., and Kaback H. R. (2014) The Life and Times of Lac Permease: Crystals Ain’t Enough, but They Certainly do Help. In Membrane transporter function: To structure and beyond (Ziegler C., and Kraemer R., Eds.) Springer Series in Biophysics: Transporters, Springer, Berlin.
-
- Abramson J.; Smirnova I.; Kasho V.; Verner G.; Kaback H. R.; Iwata S. (2003) Structure and mechanism of the lactose permease of Escherichia coli. Science 301, 610–615. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

