Overexpression of BnWRKY33 in oilseed rape enhances resistance to Sclerotinia sclerotiorum
- PMID: 24521393
- PMCID: PMC6638750
- DOI: 10.1111/mpp.12123
Overexpression of BnWRKY33 in oilseed rape enhances resistance to Sclerotinia sclerotiorum
Abstract
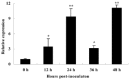
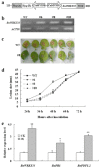
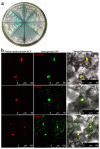
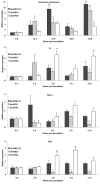
Sclerotinia sclerotiorum causes a devastating disease in oilseed rape (Brassica napus) resulting in a tremendous yield loss worldwide. Studies on various host-pathogen interactions have shown that plant WRKY transcription factors are essential for defence. For the B. napus-S. sclerotiorum interaction, little direct evidence has been found with regard to the biological roles of specific WRKY genes in host resistance. In this study, we isolated a B. napus WRKY gene, BnWRKY33, and found that the gene is highly responsive to S. sclerotiorum infection. Transgenic B. napus plants overexpressing BnWRKY33 showed markedly enhanced resistance to S. sclerotiorum, constitutive activation of the expression of BnPR1 and BnPDF1.2, and inhibition of H2 O2 accumulation in response to pathogen infection. Further, we isolated a mitogen-activated protein (MAP) kinase substrate gene, BnMKS1, and found that not only can BnWRKY33 interact with BnMKS1, which can also interact with BnMPK4, using the yeast two-hybrid assay, consistent with their collective nuclear localization, but also BnWRKY33, BnMKS1 and BnMPK4 are substantially and synergistically expressed in response to S. sclerotiorum infection. In contrast, the three genes showed differential expression in response to phytohormone treatments. Together, these results suggest that BnWRKY33 plays an important role in B. napus defence to S. sclerotiorum, which is most probably associated with the activation of the salicylic acid (SA)- and jasmonic acid (JA)-mediated defence response and inhibition of H2 O2 accumulation, and we propose a potential mechanism in which BnMPK4-BnMKS1-BnWRKY33 exist in a nuclear localized complex to regulate resistance to S. sclerotiorum in oilseed rape.
Keywords: Brassica napus; Sclerotinia sclerotiorum; WRKY33 transcription factor; disease resistance.
© 2014 BSPP AND JOHN WILEY & SONS LTD.
Figures

References
-
- Anderson, J.P. , Badruzsaufari, E. , Schenk, P.M. , Manners, J.M. , Desmond, O.J. , Ehlert, C. , Maclean, D.J. , Ebert, P.R. and Kazan, K. (2004) Antagonistic interaction between abscisic acid and jasmonate‐ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell, 16, 3460–3479. - PMC - PubMed
-
- Andreasson, E. , Jenkins, T. , Brodersen, P. , Thorgrimsen, S. , Petersen, N.H. , Zhu, S. , Qiu, J.L. , Micheelsen, P. , Rocher, A. , Petersen, M. , Newman, M.A. , Nielsen, H.B. , Hirt, H. , Somssich, I. , Mattsson, O. and Mundy, J. (2005) The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 24, 2579–2589. - PMC - PubMed
-
- Andreasson, E. , Brodersen, P. , Jenkins, T. , Mundy, J. , Petersen, N.H. , Thorgrimsen, S. and Rocher, A. (2007) Plant disease resistance and SAR regulator protein. United States Patent No.: 2007/0271623 A1.
-
- Apel, K. and Hirt, H. (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

