A systems-wide comparison of red rice (Oryza longistaminata) tissues identifies rhizome specific genes and proteins that are targets for cultivated rice improvement
- PMID: 24521476
- PMCID: PMC3933257
- DOI: 10.1186/1471-2229-14-46
A systems-wide comparison of red rice (Oryza longistaminata) tissues identifies rhizome specific genes and proteins that are targets for cultivated rice improvement
Abstract
Background: The rhizome, the original stem of land plants, enables species to invade new territory and is a critical component of perenniality, especially in grasses. Red rice (Oryza longistaminata) is a perennial wild rice species with many valuable traits that could be used to improve cultivated rice cultivars, including rhizomatousness, disease resistance and drought tolerance. Despite these features, little is known about the molecular mechanisms that contribute to rhizome growth, development and function in this plant.
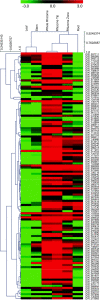
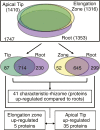
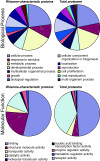
Results: We used an integrated approach to compare the transcriptome, proteome and metabolome of the rhizome to other tissues of red rice. 116 Gb of transcriptome sequence was obtained from various tissues and used to identify rhizome-specific and preferentially expressed genes, including transcription factors and hormone metabolism and stress response-related genes. Proteomics and metabolomics approaches identified 41 proteins and more than 100 primary metabolites and plant hormones with rhizome preferential accumulation. Of particular interest was the identification of a large number of gene transcripts from Magnaportha oryzae, the fungus that causes rice blast disease in cultivated rice, even though the red rice plants showed no sign of disease.
Conclusions: A significant set of genes, proteins and metabolites appear to be specifically or preferentially expressed in the rhizome of O. longistaminata. The presence of M. oryzae gene transcripts at a high level in apparently healthy plants suggests that red rice is resistant to this pathogen, and may be able to provide genes to cultivated rice that will enable resistance to rice blast disease.
Figures






References
-
- Khush G. Productivity improvements in rice. Nutr Rev. 2003;61(6 Pt 2):S114–S116. - PubMed
-
- Brar DS, Khush GS. Alien introgression in rice. Plant Mol Biol. 1997;35(1–2):35–47. - PubMed
-
- Maekawa M, Inukai T, Rikiishi K, Matsuura T, Noda K. Inheritance of the rhizomatous trait in hybrids of Oryza longistaminata Chev. et Roehr. and O. sativa L. SABRAO J Breed Genet. 1998;30:69–72.
-
- Gichuhi E, Himi E, Takahashi H, Maekawa M. Oryza longistaminata’s chromosome segments are responsible for agronomically important traits for environmentally smart rice. 2012. pp. 723–729. (Proceedings of the 2012 JKUAT Scientific, Technological and Industrialization Conference).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

