Bacteria, phages and pigs: the effects of in-feed antibiotics on the microbiome at different gut locations
- PMID: 24522263
- PMCID: PMC4817603
- DOI: 10.1038/ismej.2014.12
Bacteria, phages and pigs: the effects of in-feed antibiotics on the microbiome at different gut locations
Abstract
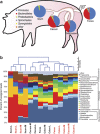
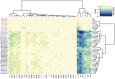
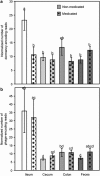
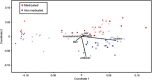
Disturbance of the beneficial gut microbial community is a potential collateral effect of antibiotics, which have many uses in animal agriculture (disease treatment or prevention and feed efficiency improvement). Understanding antibiotic effects on bacterial communities at different intestinal locations is essential to realize the full benefits and consequences of in-feed antibiotics. In this study, we defined the lumenal and mucosal bacterial communities from the small intestine (ileum) and large intestine (cecum and colon) plus feces, and characterized the effects of in-feed antibiotics (chlortetracycline, sulfamethazine and penicillin (ASP250)) on these communities. 16S rRNA gene sequence and metagenomic analyses of bacterial membership and functions revealed dramatic differences between small and large intestinal locations, including enrichment of Firmicutes and phage-encoding genes in the ileum. The large intestinal microbiota encoded numerous genes to degrade plant cell wall components, and these genes were lacking in the ileum. The mucosa-associated ileal microbiota harbored greater bacterial diversity than the lumen but similar membership to the mucosa of the large intestine, suggesting that most gut microbes can associate with the mucosa and might serve as an inoculum for the lumen. The collateral effects on the microbiota of antibiotic-fed animals caused divergence from that of control animals, with notable changes being increases in Escherichia coli populations in the ileum, Lachnobacterium spp. in all gut locations, and resistance genes to antibiotics not administered. Characterizing the differential metabolic capacities and response to perturbation at distinct intestinal locations will inform strategies to improve gut health and food safety.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

