LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma
- PMID: 24522499
- PMCID: PMC4215280
- DOI: 10.1136/gutjnl-2013-305806
LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma
Abstract
Background: Oesophageal cancer is one of the most deadly forms of cancer worldwide. Long non-coding RNAs (lncRNAs) are often found to have important regulatory roles.
Objective: To assess the lncRNA expression profile of oesophageal squamous cell carcinoma (OSCC) and identify prognosis-related lncRNAs.
Method: LncRNA expression profiles were studied by microarray in paired tumour and normal tissues from 119 patients with OSCC and validated by qRT-PCR. The 119 patients were divided randomly into training (n=60) and test (n=59) groups. A prognostic signature was developed from the training group using a random Forest supervised classification algorithm and a nearest shrunken centroid algorithm, then validated in a test group and further, in an independent cohort (n=60). The independence of the signature in survival prediction was evaluated by multivariable Cox regression analysis.
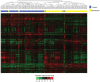
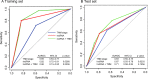
Results: LncRNAs showed significantly altered expression in OSCC tissues. From the training group, we identified a three-lncRNA signature (including the lncRNAs ENST00000435885.1, XLOC_013014 and ENST00000547963.1) which classified the patients into two groups with significantly different overall survival (median survival 19.2 months vs >60 months, p<0.0001). The signature was applied to the test group (median survival 21.5 months vs >60 months, p=0.0030) and independent cohort (median survival 25.8 months vs >48 months, p=0.0187) and showed similar prognostic values in both. Multivariable Cox regression analysis showed that the signature was an independent prognostic factor for patients with OSCC. Stratified analysis suggested that the signature was prognostic within clinical stages.
Conclusions: Our results suggest that the three-lncRNA signature is a new biomarker for the prognosis of patients with OSCC, enabling more accurate prediction of survival.
Keywords: Oesophageal Cancer.
Published by the BMJ Publishing Group Limited. For permission to use (where not already granted under a licence) please go to http://group.bmj.com/group/rights-licensing/permissions.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
