SYK allelic loss and the role of Syk-regulated genes in breast cancer survival
- PMID: 24523870
- PMCID: PMC3921124
- DOI: 10.1371/journal.pone.0087610
SYK allelic loss and the role of Syk-regulated genes in breast cancer survival
Abstract
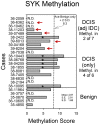
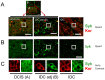
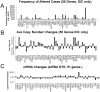
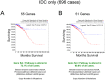
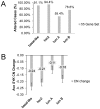
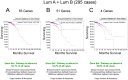
Heterozygotic loss of SYK, a non-receptor tyrosine kinase, gives rise to mouse mammary tumor formation where Syk protein levels are reduced by about half; loss of SYK mRNA is correlated with invasive cell behavior in in vitro models; and SYK loss has been correlated with distant metastases in patients. Here, allelic loss of the SYK gene was explored in breast ductal carcinoma in situ (DCIS) using fluorescence in situ hybridization and pyrosequencing, respectively, and in infiltrating ductal carcinoma (IDC) using genomic data from The Cancer Genome Atlas (TCGA). Allelic loss was present in a subset of DCIS cases where adjacent IDC was present. SYK copy number loss was found in about 26% of 1002 total breast cancer cases and 30% of IDC cases. Quantitative immunofluorescence revealed Syk protein to be six-fold higher in infiltrating immune cells compared with epithelial cells. This difference distorted tumor cell mRNA and protein levels in extracts. 20% of 1002 IDC cases contained elevated immune cell infiltration as estimated by elevated immune-specific mRNAs. In cases without immune cell infiltration, loss of SYK copy number was associated with a significant reduction of SYK mRNA. Here we define a 55 Gene Set consisting of Syk interacting, motility- and invasion-related genes. We found that overall survival was significantly reduced in IDC and Luminal A+B cases where copy number and mutations of these 55 genes were affected (Kaplan-Meier, Logrank test p-value 0.007141 and Logrank test p-value 0.001198, respectively). We conclude that reduction in Syk expression and contributions of genomic instability to copy number and mutations in the 55 Syk interacting genes significantly contribute to poorer overall patient survival. A closer examination of the role of Syk interacting motility and invasion genes and their prognostic and/or causative association with metastatic disease and patient outcome is warranted.
Conflict of interest statement
Figures





References
-
- Coopman PJ, Do MT, Barth M, Bowden ET, Hayes AJ, et al. (2000) The Syk tyrosine kinase suppresses malignant growth of human breast cancer cells. Nature 406: 742–747. - PubMed
-
- Moroni M, Soldatenkov V, Zhang L, Zhang Y, Stoica G, et al. (2004) Progressive loss of Syk and abnormal proliferation in breast cancer cells. Cancer Res 64: 7346–7354. - PubMed
-
- Coopman PJ, Mueller SC (2006) The Syk tyrosine kinase: a new negative regulator in tumor growth and progression. Cancer Letters 241: 159–173. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

