The familial Alzheimer's disease APPV717I mutation alters APP processing and Tau expression in iPSC-derived neurons
- PMID: 24524897
- PMCID: PMC4049307
- DOI: 10.1093/hmg/ddu064
The familial Alzheimer's disease APPV717I mutation alters APP processing and Tau expression in iPSC-derived neurons
Abstract
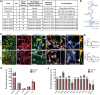
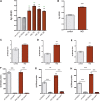
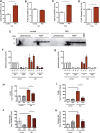
Alzheimer's disease (AD) is a complex neurodegenerative disorder characterized by extracellular plaques containing amyloid β (Aβ)-protein and intracellular tangles containing hyperphosphorylated Tau protein. Here, we describe the generation of inducible pluripotent stem cell lines from patients harboring the London familial AD (fAD) amyloid precursor protein (APP) mutation (V717I). We examine AD-relevant phenotypes following directed differentiation to forebrain neuronal fates vulnerable in AD. We observe that over differentiation time to mature neuronal fates, APP expression and levels of Aβ increase dramatically. In both immature and mature neuronal fates, the APPV717I mutation affects both β- and γ-secretase cleavage of APP. Although the mutation lies near the γ-secretase cleavage site in the transmembrane domain of APP, we find that β-secretase cleavage of APP is elevated leading to generation of increased levels of both APPsβ and Aβ. Furthermore, we find that this mutation alters the initial cleavage site of γ-secretase, resulting in an increased generation of both Aβ42 and Aβ38. In addition to altered APP processing, an increase in levels of total and phosphorylated Tau is observed in neurons with the APPV717I mutation. We show that treatment with Aβ-specific antibodies early in culture reverses the phenotype of increased total Tau levels, implicating altered Aβ production in fAD neurons in this phenotype. These studies use human neurons to reveal previously unrecognized effects of the most common fAD APP mutation and provide a model system for testing therapeutic strategies in the cell types most relevant to disease processes.
© The Author 2014. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures



References
-
- Bertram L., Lill C.M., Tanzi R.E. The genetics of Alzheimer disease: back to the future. Neuron. 2010;68:270–281. - PubMed
-
- Wolfe M.S., Xia W., Ostaszewski B.L., Diehl T.S., Kimberly W.T., Selkoe D.J. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature. 1999;398:513–517. - PubMed
-
- Selkoe D.J., Podlisny M.B. Deciphering the genetic basis of Alzheimer's disease. Annu. Rev. Genomics Hum. Genet. 2002;3:67–99. - PubMed
-
- Bertram L., Tanzi R.E. The genetics of Alzheimer's disease. Prog. Mol. Biol. Transl. Sci. 2012;107:79–100. - PubMed
-
- Goate A., Chartier-Harlin M.C., Mullan M., Brown J., Crawford F., Fidani L., Giuffra L., Haynes A., Irving N., James L. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature. 1991;349:704–706. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

