Interspecies systems biology uncovers metabolites affecting C. elegans gene expression and life history traits
- PMID: 24529378
- PMCID: PMC4169190
- DOI: 10.1016/j.cell.2014.01.047
Interspecies systems biology uncovers metabolites affecting C. elegans gene expression and life history traits
Erratum in
- Cell. 2014 Mar 13;156(6):1336-7
Abstract
Diet greatly influences gene expression and physiology. In mammals, elucidating the effects and mechanisms of individual nutrients is challenging due to the complexity of both the animal and its diet. Here, we used an interspecies systems biology approach with Caenorhabditis elegans and two of its bacterial diets, Escherichia coli and Comamonas aquatica, to identify metabolites that affect the animal's gene expression and physiology. We identify vitamin B12 as the major dilutable metabolite provided by Comamonas aq. that regulates gene expression, accelerates development, and reduces fertility but does not affect lifespan. We find that vitamin B12 has a dual role in the animal: it affects development and fertility via the methionine/S-Adenosylmethionine (SAM) cycle and breaks down the short-chain fatty acid propionic acid, preventing its toxic buildup. Our interspecies systems biology approach provides a paradigm for understanding complex interactions between diet and physiology.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
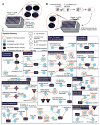
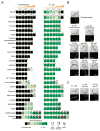


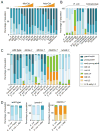
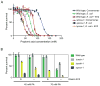
Comment in
-
Interspecies systems biology (a.k.a. interspecies genetics).Nat Methods. 2014 Apr;11(4):372. doi: 10.1038/nmeth.2917. Nat Methods. 2014. PMID: 24818228 No abstract available.
References
-
- Al-Lahham SH, Peppelenbosch MP, Roelofsen H, Vonk RJ, Venema K. Biological effects of propionic acid in humans; metabolism, potential applications and underlying mechanisms. Biochimica et biophysica acta. 2010;1801:1175–1183. - PubMed
-
- Albert MJ, Mathan VI, Baker SJ. Vitamin B12 synthesis by human small intestinal bacteria. Nature. 1980;283:781–782. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

