Application of activity-based protein profiling to study enzyme function in adipocytes
- PMID: 24529438
- PMCID: PMC4138146
- DOI: 10.1016/B978-0-12-800280-3.00009-8
Application of activity-based protein profiling to study enzyme function in adipocytes
Abstract
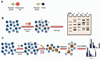
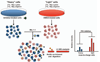
Activity-based protein profiling (ABPP) is a chemical proteomics approach that utilizes small-molecule probes to determine the functional state of enzymes directly in native systems. ABPP probes selectively label active enzymes, but not their inactive forms, facilitating the characterization of changes in enzyme activity that occur without alterations in protein levels. ABPP can be a tool superior to conventional gene expression and proteomic profiling methods to discover new enzymes active in adipocytes and to detect differences in the activity of characterized enzymes that may be associated with disorders of adipose tissue function. ABPP probes have been developed that react selectively with most members of specific enzyme classes. Here, using as an example the serine hydrolase family that includes many enzymes with critical roles in adipocyte physiology, we describe methods to apply ABPP analysis to the study of adipocyte enzymatic pathways.
Keywords: ABPP probes; Activity-based protein profiling; Adipocyte enzymes; Chemoproteomics methods; Enzyme activity; Functional proteomics; Serine hydrolases.
© 2014 Elsevier Inc. All rights reserved.
Figures



References
-
- Adam GC, Sorensen EJ, Cravatt BF. Proteomic profiling of mechanistically distinct enzyme classes using a common chemotype. Nature Biotechnology. 2002;20:805–809. - PubMed
-
- Adibekian A, Martin BR, Chang JW, Hsu KL, Tsuboi K, Bachovchin DA, Speers AE, Brown SJ, Spicer T, Fernandez-Vega V, Ferguson J, Hodder PS, Rosen H, Cravatt BF. Confirming target engagement for reversible inhibitors in vivo by kinetically tuned activity-based probes. J Am Chem Soc. 2012;134(25):10345–10348. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

