Molecular and transcriptional basis of CD4⁺ T cell dysfunction during chronic infection
- PMID: 24530057
- PMCID: PMC3990591
- DOI: 10.1016/j.immuni.2014.01.005
Molecular and transcriptional basis of CD4⁺ T cell dysfunction during chronic infection
Abstract
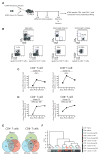
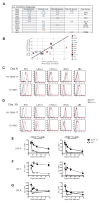
T cell exhaustion is common during chronic infections. Although CD4(+) T cells are critical for controlling viral load during chronic viral infections, less is known about their differentiation and transcriptional program. We defined the phenotypic, functional, and molecular profiles of exhausted CD4(+) T cells. Global transcriptional analysis demonstrated a molecular profile distinct from effector and memory CD4(+) T cells and also from exhausted CD8(+) T cells, though some common features of CD4(+) and CD8(+) T cell exhaustion were revealed. We have demonstrated unappreciated roles for transcription factors (TFs) including Helios, type I interferon (IFN-I) signaling, and a diverse set of coinhibitory and costimulatory molecules during CD4(+) T cell exhaustion. Moreover, the signature of CD4(+) T cell exhaustion was found to be distinct from that of other CD4(+) T cell lineage subsets and was associated with TF heterogeneity. This study provides a framework for therapeutic interventions targeting exhausted CD4(+) T cells.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures





References
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

