Sparse representation based biomarker selection for schizophrenia with integrated analysis of fMRI and SNPs
- PMID: 24530838
- PMCID: PMC4130811
- DOI: 10.1016/j.neuroimage.2014.01.021
Sparse representation based biomarker selection for schizophrenia with integrated analysis of fMRI and SNPs
Abstract
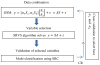
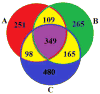
Integrative analysis of multiple data types can take advantage of their complementary information and therefore may provide higher power to identify potential biomarkers that would be missed using individual data analysis. Due to different natures of diverse data modality, data integration is challenging. Here we address the data integration problem by developing a generalized sparse model (GSM) using weighting factors to integrate multi-modality data for biomarker selection. As an example, we applied the GSM model to a joint analysis of two types of schizophrenia data sets: 759,075 SNPs and 153,594 functional magnetic resonance imaging (fMRI) voxels in 208 subjects (92 cases/116 controls). To solve this small-sample-large-variable problem, we developed a novel sparse representation based variable selection (SRVS) algorithm, with the primary aim to identify biomarkers associated with schizophrenia. To validate the effectiveness of the selected variables, we performed multivariate classification followed by a ten-fold cross validation. We compared our proposed SRVS algorithm with an earlier sparse model based variable selection algorithm for integrated analysis. In addition, we compared with the traditional statistics method for uni-variant data analysis (Chi-squared test for SNP data and ANOVA for fMRI data). Results showed that our proposed SRVS method can identify novel biomarkers that show stronger capability in distinguishing schizophrenia patients from healthy controls. Moreover, better classification ratios were achieved using biomarkers from both types of data, suggesting the importance of integrative analysis.
Keywords: SNP; Schizophrenia; Sparse representations; Variable selection; fMRI.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures





References
-
- Badner JA, Gershon ES. Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol Psychiatry. 2002;7(4):405–411. - PubMed
-
- Cai TT, Wang L. Orthogonal Matching Pursuit for Sparse Signal Recovery. IEEE Trans on Inf Theory. 2011;57(7):1–26.
-
- Callicott JH, Straub RE, Pezawas L, Egan MF, Mattay VS, Hariri AR, Verchinski BA, Meyer-Lindenberg A, Balkissoon R, Kolachana B, Goldberg TE, Weinberger DR. Variation in DISC1 affects hippocampal structure and function and increases risk for schizophrenia. Proc Natl Acad Sci U S A. 2005 Jun;102(24):8627–32. - PMC - PubMed
-
- Candès E, Tao T. Near optimal signal recovery from random projections:Universal encoding strategies? IEEE Trans Inf Theory Dec. 2006;52(12):5406–5425.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

