The spliceosome: disorder and dynamics defined
- PMID: 24530854
- PMCID: PMC3987960
- DOI: 10.1016/j.sbi.2014.01.009
The spliceosome: disorder and dynamics defined
Abstract
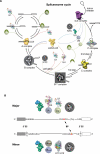
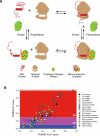
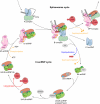
Among the many macromolecular machines involved in eukaryotic gene expression, the spliceosome remains one of the most challenging for structural biologists. Defining features of this highly complex apparatus are its excessive number of individual parts, many of which have been evolutionarily selected for regions of structural disorder, and the remarkable compositional and conformation dynamics it must undertake to complete each round of splicing. Here we review recent advances in our understanding of spliceosome structural dynamics stemming from bioinformatics, deep sequencing, high throughput methods for determining protein-protein, protein-RNA and RNA-RNA interaction dynamics, single molecule microscopy and more traditional structural analyses. Together, these tools are rapidly changing our structural appreciation of this remarkably dynamic machine.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures
References
-
- Bhattacharya A. Protein structures: Structures of desire. Nature. 2009;459:24–27. - PubMed
-
- Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends in Genetics. 2001;17:100–107. - PubMed
-
- Nagai K. The upcoming review, Current Opinion in Structural Biology. in press.
-
- Jurica MS, Moore MJ. Pre-mRNA Splicing: Awash in a Sea of Proteins. Molecular Cell. 2003;12:5–14. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

