CRISPR adaptive immune systems of Archaea
- PMID: 24531374
- PMCID: PMC3973734
- DOI: 10.4161/rna.27990
CRISPR adaptive immune systems of Archaea
Abstract
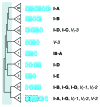
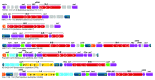
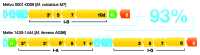
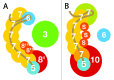
CRISPR adaptive immune systems were analyzed for all available completed genomes of archaea, which included representatives of each of the main archaeal phyla. Initially, all proteins encoded within, and proximal to, CRISPR-cas loci were clustered and analyzed using a profile-profile approach. Then cas genes were assigned to gene cassettes and to functional modules for adaptation and interference. CRISPR systems were then classified primarily on the basis of their concatenated Cas protein sequences and gene synteny of the interference modules. With few exceptions, they could be assigned to the universal Type I or Type III systems. For Type I, subtypes I-A, I-B, and I-D dominate but the data support the division of subtype I-B into two subtypes, designated I-B and I-G. About 70% of the Type III systems fall into the universal subtypes III-A and III-B but the remainder, some of which are phyla-specific, diverge significantly in Cas protein sequences, and/or gene synteny, and they are classified separately. Furthermore, a few CRISPR systems that could not be assigned to Type I or Type III are categorized as variant systems. Criteria are presented for assigning newly sequenced archaeal CRISPR systems to the different subtypes. Several accessory proteins were identified that show a specific gene linkage, especially to Type III interference modules, and these may be cofunctional with the CRISPR systems. Evidence is presented for extensive exchange having occurred between adaptation and interference modules of different archaeal CRISPR systems, indicating the wide compatibility of the functionally diverse interference complexes with the relatively conserved adaptation modules.
Keywords: CRISPR; Cas; Type I; Type III; archaea.
Figures



References
-
- Barrangou R, van der Oost J, eds. CRISPR-Cas systems. Springer press, Heidelberg, 2012; pp. 1-299.
-
- Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
