Sequencing, assembling, and correcting draft genomes using recombinant populations
- PMID: 24531727
- PMCID: PMC4059239
- DOI: 10.1534/g3.114.010264
Sequencing, assembling, and correcting draft genomes using recombinant populations
Abstract
Current de novo whole-genome sequencing approaches often are inadequate for organisms lacking substantial preexisting genetic data. Problems with these methods are manifest as: large numbers of scaffolds that are not ordered within chromosomes or assigned to individual chromosomes, misassembly of allelic sequences as separate loci when the individual(s) being sequenced are heterozygous, and the collapse of recently duplicated sequences into a single locus, regardless of levels of heterozygosity. Here we propose a new approach for producing de novo whole-genome sequences-which we call recombinant population genome construction-that solves many of the problems encountered in standard genome assembly and that can be applied in model and nonmodel organisms. Our approach takes advantage of next-generation sequencing technologies to simultaneously barcode and sequence a large number of individuals from a recombinant population. The sequences of all recombinants can be combined to create an initial de novo assembly, followed by the use of individual recombinant genotypes to correct assembly splitting/collapsing and to order and orient scaffolds within linkage groups. Recombinant population genome construction can rapidly accelerate the transformation of nonmodel species into genome-enabled systems by simultaneously producing a high-quality genome assembly and providing genomic tools (e.g., high-confidence single-nucleotide polymorphisms) for immediate applications. In populations segregating for important functional traits, this approach also enables simultaneous mapping of quantitative trait loci. We demonstrate our method using simulated Illumina data from a recombinant population of Caenorhabditis elegans and show that the method can produce a high-fidelity, high-quality genome assembly for both parents of the cross.
Keywords: assembly; duplication; genetics; genome; next-generation sequencing.
Figures

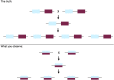
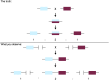
References
-
- Adams M. D., Celniker S. E., Holt R. A., Evans C. A., Gocayne J. D., et al. , 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. - PubMed
-
- Bailey J. A., Gu Z. P., Clark R. A., Reinert K., Samonte R. V., et al. , 2002. Recent segmental duplications in the human genome. Science 297: 1003–1007. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
