TcaR-ssDNA complex crystal structure reveals new DNA binding mechanism of the MarR family proteins
- PMID: 24531929
- PMCID: PMC4005659
- DOI: 10.1093/nar/gku128
TcaR-ssDNA complex crystal structure reveals new DNA binding mechanism of the MarR family proteins
Abstract
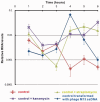
The teicoplanin-associated locus regulator (TcaR) regulates gene expression of proteins on the intercellular adhesion (ica) locus involved in staphylococci poly-N-acetylglucosamine biosynthesis. The absence of TcaR increases poly-N-acetylglucosamine production and promotes biofilm formation. Until recently, the mechanism of multiple antibiotic resistance regulator family protein members, such as TcaR, was restricted to binding double-stranded DNA. However, we recently found that TcaR strongly interacts with single-stranded DNA, which is a new role for this family of proteins. In this study, we report Staphylococcus epidermidis TcaR-single-stranded DNA complex structures. Our model suggests that TcaR and single-stranded DNA form a 61-symmetry polymer composed of TcaR dimers with single-stranded DNA that wraps outside the polymer and 12 nt per TcaR dimer. Single-stranded DNA binding to TcaR involves a large conformational change at the DNA binding lobe. Several point mutations involving the single-stranded DNA binding surface validate interactions between single-stranded DNA and TcaR. Our results extend the novel role of multiple antibiotic resistance regulator family proteins in staphylococci.
Figures


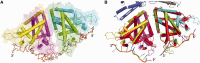

References
-
- Anderson SD, Gums JG. Ceftobiprole: an extended-spectrum anti-methicillin-resistant Staphylococcus aureus cephalosporin. Ann. Pharmacother. 2008;42:806–816. - PubMed
-
- Brandenberger M, Tschierske M, Giachino P, Wada A, Berger-Bachi B. Inactivation of a novel three-cistronic operon tcaR-tcaA-tcaB increases teicoplanin resistance in Staphylococcus aureus. Biochim. Biophys. Acta. 2000;1523:135–139. - PubMed
-
- Tegmark K, Karlsson A, Arvidson S. Identification and characterization of SarH1: a new global regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 2000;37:398–409. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

