Microbial species diversity, community dynamics, and metabolite kinetics of water kefir fermentation
- PMID: 24532061
- PMCID: PMC3993195
- DOI: 10.1128/AEM.03978-13
Microbial species diversity, community dynamics, and metabolite kinetics of water kefir fermentation
Abstract
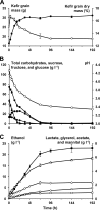
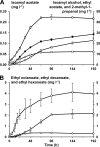
Water kefir is a sour, alcoholic, and fruity fermented beverage of which the fermentation is started with water kefir grains. These water kefir grains consist of polysaccharide and contain the microorganisms responsible for the water kefir fermentation. In this work, a water kefir fermentation process was followed as a function of time during 192 h to unravel the community dynamics, the species diversity, and the kinetics of substrate consumption and metabolite production. The majority of the water kefir ecosystem was found to be present on the water kefir grains. The most important microbial species present were Lactobacillus casei/paracasei, Lactobacillus harbinensis, Lactobacillus hilgardii, Bifidobacterium psychraerophilum/crudilactis, Saccharomyces cerevisiae, and Dekkera bruxellensis. The microbial species diversities in the water kefir liquor and on the water kefir grains were similar and remained stable during the whole fermentation process. The major substrate, sucrose, was completely converted after 24 h of fermentation, which coincided with the production of the major part of the water kefir grain polysaccharide. The main metabolites of the fermentation were ethanol and lactic acid. Glycerol, acetic acid, and mannitol were produced in low concentrations. The major part of these metabolites was produced during the first 72 h of fermentation, during which the pH decreased from 4.26 to 3.45. The most prevalent volatile aroma compounds were ethyl acetate, isoamyl acetate, ethyl hexanoate, ethyl octanoate, and ethyl decanoate, which might be of significance with respect to the aroma of the end product.
Figures



Similar articles
-
Shotgun Metagenomics of a Water Kefir Fermentation Ecosystem Reveals a Novel Oenococcus Species.Front Microbiol. 2019 Mar 13;10:479. doi: 10.3389/fmicb.2019.00479. eCollection 2019. Front Microbiol. 2019. PMID: 30918501 Free PMC article.
-
The Buffer Capacity and Calcium Concentration of Water Influence the Microbial Species Diversity, Grain Growth, and Metabolite Production During Water Kefir Fermentation.Front Microbiol. 2019 Dec 13;10:2876. doi: 10.3389/fmicb.2019.02876. eCollection 2019. Front Microbiol. 2019. PMID: 31921054 Free PMC article.
-
The Type and Concentration of Inoculum and Substrate as Well as the Presence of Oxygen Impact the Water Kefir Fermentation Process.Front Microbiol. 2021 Feb 11;12:628599. doi: 10.3389/fmicb.2021.628599. eCollection 2021. Front Microbiol. 2021. PMID: 33643256 Free PMC article.
-
An update on water kefir: Microbiology, composition and production.Int J Food Microbiol. 2021 May 2;345:109128. doi: 10.1016/j.ijfoodmicro.2021.109128. Epub 2021 Mar 3. Int J Food Microbiol. 2021. PMID: 33751986 Review.
-
The effect of kefir produced from natural kefir grains on the intestinal microbial populations and antioxidant capacities of Balb/c mice.Food Res Int. 2019 Jan;115:408-413. doi: 10.1016/j.foodres.2018.10.080. Epub 2018 Oct 30. Food Res Int. 2019. PMID: 30599959 Review.
Cited by
-
Wort Substrate Consumption and Metabolite Production During Lambic Beer Fermentation and Maturation Explain the Successive Growth of Specific Bacterial and Yeast Species.Front Microbiol. 2018 Nov 19;9:2763. doi: 10.3389/fmicb.2018.02763. eCollection 2018. Front Microbiol. 2018. PMID: 30510547 Free PMC article.
-
Microbial viability and nutritional content of water kefir grains under different storage conditions.Food Sci Nutr. 2024 Mar 5;12(6):4143-4150. doi: 10.1002/fsn3.4074. eCollection 2024 Jun. Food Sci Nutr. 2024. PMID: 38873456 Free PMC article.
-
Technological and Environmental Features Determine the Uniqueness of the Lambic Beer Microbiota and Production Process.Appl Environ Microbiol. 2021 Aug 26;87(18):e0061221. doi: 10.1128/AEM.00612-21. Epub 2021 Aug 26. Appl Environ Microbiol. 2021. PMID: 34232060 Free PMC article. Review.
-
Kefir and Its Biological Activities.Foods. 2021 May 27;10(6):1210. doi: 10.3390/foods10061210. Foods. 2021. PMID: 34071977 Free PMC article. Review.
-
Strategies to Improve the Potential Functionality of Fruit-Based Fermented Beverages.Plants (Basel). 2021 Oct 22;10(11):2263. doi: 10.3390/plants10112263. Plants (Basel). 2021. PMID: 34834623 Free PMC article. Review.
References
-
- Pidoux M. 1989. The microbial flora of sugary kefir grain (the gingerbeer plant) - biosynthesis of the grain from Lactobacillus hilgardii producing a polysaccharide gel. J. Appl. Microbiol. 5:223–238. 10.1007/Bf01741847 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

