Characterization of temperate phages infecting Clostridium difficile isolates of human and animal origins
- PMID: 24532062
- PMCID: PMC3993186
- DOI: 10.1128/AEM.00237-14
Characterization of temperate phages infecting Clostridium difficile isolates of human and animal origins
Abstract
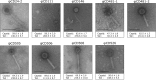
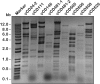
Clostridium difficile is a Gram-positive pathogen infecting humans and animals. Recent studies suggest that animals could represent potential reservoirs of C. difficile that could then transfer to humans. Temperate phages contribute to the evolution of most bacteria, for example, by promoting the transduction of virulence, fitness, and antibiotic resistance genes. In C. difficile, little is known about their role, mainly because suitable propagating hosts and conditions are lacking. Here we report the isolation, propagation, and preliminary characterization of nine temperate phages from animal and human C. difficile isolates. Prophages were induced by UV light from 58 C. difficile isolates of animal and human origins. Using soft agar overlays with 27 different C. difficile test strains, we isolated and further propagated nine temperate phages: two from horse isolates (ΦCD481-1 and ΦCD481-2), three from dog isolates (ΦCD505, ΦCD506, and ΦCD508), and four from human isolates (ΦCD24-2, ΦCD111, ΦCD146, and ΦCD526). Two phages are members of the Siphoviridae family (ΦCD111 and ΦCD146), while the others are Myoviridae phages. Pulsed-field gel electrophoresis and restriction enzyme analyses showed that all of the phages had unique double-stranded DNA genomes of 30 to 60 kb. Phages induced from human C. difficile isolates, especially the members of the Siphoviridae family, had a broader host range than phages from animal C. difficile isolates. Nevertheless, most of the phages could infect both human and animal strains. Phage transduction of antibiotic resistance was recently reported in C. difficile. Our findings therefore call for further investigation of the potential risk of transduction between animal and human C. difficile isolates.
Figures

Similar articles
-
Bacteriophages Contribute to Shaping Clostridioides (Clostridium) difficile Species.Front Microbiol. 2018 Aug 31;9:2033. doi: 10.3389/fmicb.2018.02033. eCollection 2018. Front Microbiol. 2018. PMID: 30233520 Free PMC article. Review.
-
Characterization of Functional Prophages in Clostridium difficile.Methods Mol Biol. 2016;1476:143-65. doi: 10.1007/978-1-4939-6361-4_11. Methods Mol Biol. 2016. PMID: 27507339
-
Prophage carriage and diversity within clinically relevant strains of Clostridium difficile.Appl Environ Microbiol. 2012 Sep;78(17):6027-34. doi: 10.1128/AEM.01311-12. Epub 2012 Jun 15. Appl Environ Microbiol. 2012. PMID: 22706062 Free PMC article.
-
Prophage-stimulated toxin production in Clostridium difficile NAP1/027 lysogens.J Bacteriol. 2011 Jun;193(11):2726-34. doi: 10.1128/JB.00787-10. Epub 2011 Mar 25. J Bacteriol. 2011. PMID: 21441508 Free PMC article.
-
Diversity, Dynamics and Therapeutic Application of Clostridioides difficile Bacteriophages.Viruses. 2022 Dec 12;14(12):2772. doi: 10.3390/v14122772. Viruses. 2022. PMID: 36560776 Free PMC article. Review.
Cited by
-
Metagenomic-based characterization of the gut virome in patients with polycystic ovary syndrome.Front Microbiol. 2022 Aug 16;13:951782. doi: 10.3389/fmicb.2022.951782. eCollection 2022. Front Microbiol. 2022. PMID: 36051758 Free PMC article.
-
Phages in Anaerobic Systems.Viruses. 2020 Sep 26;12(10):1091. doi: 10.3390/v12101091. Viruses. 2020. PMID: 32993161 Free PMC article. Review.
-
Emergence of an outbreak-associated Clostridium difficile variant with increased virulence.J Clin Microbiol. 2015 Apr;53(4):1216-26. doi: 10.1128/JCM.03058-14. Epub 2015 Feb 4. J Clin Microbiol. 2015. PMID: 25653402 Free PMC article.
-
Genetic and phenotypic characteristics of Clostridium (Clostridioides) difficile from canine, bovine, and pediatric populations.Anaerobe. 2022 Apr;74:102539. doi: 10.1016/j.anaerobe.2022.102539. Epub 2022 Feb 23. Anaerobe. 2022. PMID: 35217150 Free PMC article.
-
Bacteriophages Contribute to Shaping Clostridioides (Clostridium) difficile Species.Front Microbiol. 2018 Aug 31;9:2033. doi: 10.3389/fmicb.2018.02033. eCollection 2018. Front Microbiol. 2018. PMID: 30233520 Free PMC article. Review.
References
-
- He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, Martin MJ, Connor TR, Harris SR, Fairley D, Bamford KB, D'Arc S, Brazier J, Brown D, Coia JE, Douce G, Gerding D, Kim HJ, Koh TH, Kato H, Senoh M, Louie T, Michell S, Butt E, Peacock SJ, Brown NM, Riley T, Songer G, Wilcox M, Pirmohamed M, Kuijper E, Hawkey P, Wren BW, Dougan G, Parkhill J, Lawley TD. 2013. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat. Genet. 45:109–113. 10.1038/ng.2478 - DOI - PMC - PubMed
-
- Loo V, Poirier L, Miller M, Oughton M, Libman M, Michaud S, Bourgault A, Nguyen T, Frenette C, Kelly M, Vibien A, Brassard P, Fenn S, Dewar K, Hudson T, Horn R, Rene P, Monczak Y, Dascal A. 2005. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N. Engl. J. Med. 353:2442–2449. 10.1056/NEJMoa051639 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

