TiO2 photocatalysis damages lipids and proteins in Escherichia coli
- PMID: 24532071
- PMCID: PMC3993174
- DOI: 10.1128/AEM.03995-13
TiO2 photocatalysis damages lipids and proteins in Escherichia coli
Abstract
This study investigates the mechanisms of UV-A (315 to 400 nm) photocatalysis with titanium dioxide (TiO2) applied to the degradation of Escherichia coli and their effects on two key cellular components: lipids and proteins. The impact of TiO2 photocatalysis on E. coli survival was monitored by counting on agar plate and by assessing lipid peroxidation and performing proteomic analysis. We observed through malondialdehyde quantification that lipid peroxidation occurred during the photocatalytic process, and the addition of superoxide dismutase, which acts as a scavenger of the superoxide anion radical (O2·(-)), inhibited this effect by half, showing us that O2·(-) radicals participate in the photocatalytic antimicrobial effect. Qualitative analysis using two-dimensional electrophoresis allowed selection of proteins for which spot modifications were observed during the applied treatments. Two-dimensional electrophoresis highlighted that among the selected protein spots, 7 and 19 spots had already disappeared in the dark in the presence of 0.1 g/liter and 0.4 g/liter TiO2, respectively, which is accounted for by the cytotoxic effect of TiO2. Exposure to 30 min of UV-A radiation in the presence of 0.1 g/liter and 0.4 g/liter TiO2 increased the numbers of missing spots to 14 and 22, respectively. The proteins affected by photocatalytic oxidation were strongly heterogeneous in terms of location and functional category. We identified several porins, proteins implicated in stress response, in transport, and in bacterial metabolism. This study reveals the simultaneous effects of O2·(-) on lipid peroxidation and on the proteome during photocatalytic treatment and therefore contributes to a better understanding of molecular mechanisms in antibacterial photocatalytic treatment.
Figures

 , in the dark;
, in the dark;  , under UV-A irradiation. * (P < 0.05), ** (P < 0.01), *** (P < 0.001), and **** (P < 0.0001) indicate the significance of the difference between sample means, obtained using Student's t test.
, under UV-A irradiation. * (P < 0.05), ** (P < 0.01), *** (P < 0.001), and **** (P < 0.0001) indicate the significance of the difference between sample means, obtained using Student's t test.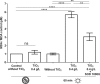
 , in the dark;
, in the dark;  , under UV-A irradiation. ** (P < 0.01), **** (P < 0.0001), and ns (not significant) indicate the significance of the difference between sample means obtained with Student's t test.
, under UV-A irradiation. ** (P < 0.01), **** (P < 0.0001), and ns (not significant) indicate the significance of the difference between sample means obtained with Student's t test.
References
-
- Wolska KI, Grzes K, Kurek A. 2012. Synergy between novel antimicrobials and conventional antibiotics or bacteriocins. Pol. J. Microbiol. 61:95–104 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

