Protein methylation and interaction with the antiproliferative gene, BTG2/TIS21/Pc3
- PMID: 24532495
- PMCID: PMC3936616
- DOI: 10.3349/ymj.2014.55.2.292
Protein methylation and interaction with the antiproliferative gene, BTG2/TIS21/Pc3
Abstract
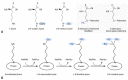
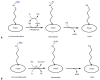
The last one and half a decade witnessed an outstanding re-emergence of attention and remarkable progress in the field of protein methylation. In the present article, we describe the early discoveries in research and review the role protein methylation played in the biological function of the antiproliferative gene, BTG2/TIS21/PC3.
Keywords: APRO (antiproliferative) genes; BTG2 (B-cell translocation gene 2); PKMT; PRMT; Protein methylation; TIS21 (TPA-inducible sequences 21); protein methyltransferases.
Conflict of interest statement
The authors have no financial conflicts of interest.
Figures


Similar articles
-
The gene PC3(TIS21/BTG2), prototype member of the PC3/BTG/TOB family: regulator in control of cell growth, differentiation, and DNA repair?J Cell Physiol. 2001 May;187(2):155-65. doi: 10.1002/jcp.1062. J Cell Physiol. 2001. PMID: 11267995 Review.
-
TIS21(/BTG2/PC3) accelerates the repair of DNA double strand breaks by enhancing Mre11 methylation and blocking damage signal transfer to the Chk2(T68)-p53(S20) pathway.DNA Repair (Amst). 2012 Dec 1;11(12):965-75. doi: 10.1016/j.dnarep.2012.09.009. Epub 2012 Oct 22. DNA Repair (Amst). 2012. PMID: 23089312
-
Regulation of Btg2(/TIS21/PC3) expression via reactive oxygen species-protein kinase C-ΝFκΒ pathway under stress conditions.Cell Signal. 2013 Dec;25(12):2400-12. doi: 10.1016/j.cellsig.2013.07.015. Epub 2013 Jul 19. Cell Signal. 2013. PMID: 23876794
-
B cell translocation gene 2 enhances susceptibility of HeLa cells to doxorubicin-induced oxidative damage.J Biol Chem. 2008 Nov 28;283(48):33110-8. doi: 10.1074/jbc.M804255200. Epub 2008 Oct 7. J Biol Chem. 2008. PMID: 18840609 Free PMC article.
-
BTG2: a rising star of tumor suppressors (review).Int J Oncol. 2015 Feb;46(2):459-64. doi: 10.3892/ijo.2014.2765. Epub 2014 Nov 18. Int J Oncol. 2015. PMID: 25405282 Review.
Cited by
-
High-throughput computational investigation of protein electrostatics and cavity for SAM-dependent methyltransferases.Protein Sci. 2023 Jul;32(7):e4690. doi: 10.1002/pro.4690. Protein Sci. 2023. PMID: 37278582 Free PMC article.
-
Control of the Cell Cycle in Adult Neurogenesis and its Relation with Physical Exercise.Brain Plast. 2015 Oct 9;1(1):41-54. doi: 10.3233/BPL-150013. Brain Plast. 2015. PMID: 29765834 Free PMC article. Review.
-
[Effect of sodium phenylbutyrate on the sensitivity of PC3/DTX-resistant prostate cancer cells to docetaxel].Nan Fang Yi Ke Da Xue Xue Bao. 2017 Novemer 20;37(1):130-134. doi: 10.3969/j.issn.1673-4254.2017.01.24. Nan Fang Yi Ke Da Xue Xue Bao. 2017. PMID: 28109113 Free PMC article. Chinese.
References
-
- Kim S, Paik WK. Studies on the origin of epsilon-N-methyl-L-lysine in protein. J Biol Chem. 1965;240:4629–4634. - PubMed
-
- Schoenheimer R, Ratner S, Rittenberg D. Studies on protein metabolism: VII. The metabolism of tyrosine. J Biol Chem. 1939;127:333–344.
-
- Ambler RP, Rees MW. Epsilon-N-methyl-lysine in bacterial flagellar protein. Nature. 1959;184:56–57. - PubMed
-
- Murray K. The occurence of epsilon-N-methyl lysine in histones. Biochemistry. 1964;3:10–15. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

