Bacteroides isolated from four mammalian hosts lack host-specific 16S rRNA gene phylogeny and carbon and nitrogen utilization patterns
- PMID: 24532571
- PMCID: PMC3996570
- DOI: 10.1002/mbo3.159
Bacteroides isolated from four mammalian hosts lack host-specific 16S rRNA gene phylogeny and carbon and nitrogen utilization patterns
Abstract
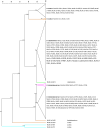
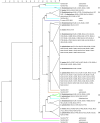
One-hundred-and-three isolates of Bacteroides ovatus, B. thetaiotaomicron, and B. xylanisolvens were recovered from cow, goat, human, and pig fecal enrichments with cellulose or xylan/pectin. Isolates were compared using 16S rRNA gene sequencing, repetitive sequence-based polymerase chain reaction (rep-PCR), and phenotypic microarrays. Analysis of 16S rRNA gene sequences revealed high sequence identity in these Bacteroides; with distinct phylogenetic groupings by bacterial species but not host origin. Phenotypic microarray analysis demonstrated these Bacteroides shared the ability to utilize many of the same carbon substrates, without differences due to species or host origin, indicative of their broad carbohydrate fermentation abilities. Limited nitrogen substrates were utilized; in addition to ammonia, guanine, and xanthine, purine derivatives were utilized by most isolates followed by a few amino sugars. Only rep-PCR analysis demonstrated host-specific patterns, indicating that genomic changes due to coevolution with host did not occur by mutation in the 16S rRNA gene or by a gain or loss of carbohydrate utilization genes within these Bacteroides. This is the first report to indicate that host-associated genomic differences are outside of 16S rRNA gene and carbohydrate utilization genes and suggest conservation of specific bacterial species with the same functionality across mammalian hosts for this Bacteroidetes clade.
Keywords: Bacteroides; cow; fecal bacteria; goat; gut microbiota; human; phenotype array; pig.
© 2014 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Figures



References
-
- Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science (New York, N. Y.) 2005;307:1915–1920. - PubMed
-
- Bergman EN. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 1990;70:567–590. - PubMed
-
- Borglin S, Joyner D, Jacobsen J, Mukhopadhyay A, Hazen TC. Overcoming the anaerobic hurdle in phenotypic microarrays: generation and visualization of growth curve data for Desulfovibrio vulgaris Hildenborough. J. Microbiol. Methods. 2009;76:159–168. - PubMed
-
- Comstock LE, Coyne MJ. Bacteroides thetaiotaomicron: a dynamic, niche-adapted human symbiont. BioEssays. 2003;25:926–929. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

