Real-Time Protein Crystallization Image Acquisition and Classification System
- PMID: 24532991
- PMCID: PMC3921687
- DOI: 10.1021/cg3016029
Real-Time Protein Crystallization Image Acquisition and Classification System
Abstract
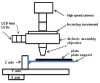
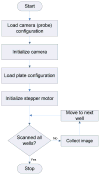
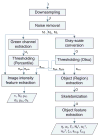
In this paper, we describe the design and implementation of a stand-alone real-time system for protein crystallization image acquisition and classification with a goal to assist crystallographers in scoring crystallization trials. In-house assembled fluorescence microscopy system is built for image acquisition. The images are classified into three categories as non-crystals, likely leads, and crystals. Image classification consists of two main steps - image feature extraction and application of classification based on multilayer perceptron (MLP) neural networks. Our feature extraction involves applying multiple thresholding techniques, identifying high intensity regions (blobs), and generating intensity and blob features to obtain a 45-dimensional feature vector per image. To reduce the risk of missing crystals, we introduce a max-class ensemble classifier which applies multiple classifiers and chooses the highest score (or class). We performed our experiments on 2250 images consisting 67% non-crystal, 18% likely leads, and 15% clear crystal images and tested our results using 10-fold cross validation. Our results demonstrate that the method is very efficient (< 3 seconds to process and classify an image) and has comparatively high accuracy. Our system only misses 1.2% of the crystals (classified as non-crystals) most likely due to low illumination or out of focus image capture and has an overall accuracy of 88%.
Figures




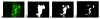



References
-
- Bern M, et al. Automatic classification of protein crystallization images using a curve-tracking algorithm. J Appl Cryst. 2004;37:279–287.
-
- Berry IM, et al. SPINE high-throughput crystallization, crystal imaging and recognition techniques: current state, performance analysis, new technologies and future aspects. Acta Cryst. 2006;D62:1137–1149. - PubMed
-
- Cumbaa CA, et al. Automatic classification of sub-microlitre protein-crystallization trials in 1536-well plates. Acta Cryst. 2003;D59:1619–1627. - PubMed
-
- Cumbaa CA, Jurisica I. Automatic classification and pattern discovery in high-throughput protein crystallization trials. J Struct Funct Gen. 2005;6:195–202. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
