Genome-wide association and systems genetic analyses of residual feed intake, daily feed consumption, backfat and weight gain in pigs
- PMID: 24533460
- PMCID: PMC3929553
- DOI: 10.1186/1471-2156-15-27
Genome-wide association and systems genetic analyses of residual feed intake, daily feed consumption, backfat and weight gain in pigs
Abstract
Background: Feed efficiency is one of the major components determining costs of animal production. Residual feed intake (RFI) is defined as the difference between the observed and the expected feed intake given a certain production. Residual feed intake 1 (RFI1) was calculated based on regression of individual daily feed intake (DFI) on initial test weight and average daily gain. Residual feed intake 2 (RFI2) was as RFI1 except it was also regressed with respect to backfat (BF). It has been shown to be a sensitive and accurate measure for feed efficiency in livestock but knowledge of the genomic regions and mechanisms affecting RFI in pigs is lacking. The study aimed to identify genetic markers and candidate genes for RFI and its component traits as well as pathways associated with RFI in Danish Duroc boars by genome-wide associations and systems genetic analyses.
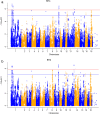
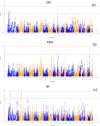
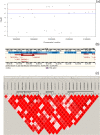
Results: Phenotypic and genotypic records (using the Illumina Porcine SNP60 BeadChip) were available on 1,272 boars. Fifteen and 12 loci were significantly associated (p < 1.52 × 10-6) with RFI1 and RFI2, respectively. Among them, 10 SNPs were significantly associated with both RFI1 and RFI2 implying the existence of common mechanisms controlling the two RFI measures. Significant QTL regions for component traits of RFI (DFI and BF) were detected on pig chromosome (SSC) 1 (for DFI) and 2 for (BF). The SNPs within MAP3K5 and PEX7 on SSC 1, ENSSSCG00000022338 on SSC 9, and DSCAM on SSC 13 might be interesting markers for both RFI measures. Functional annotation of genes in 0.5 Mb size flanking significant SNPs indicated regulation of protein and lipid metabolic process, gap junction, inositol phosphate metabolism and insulin signaling pathway are significant biological processes and pathways for RFI, respectively.
Conclusions: The study detected novel genetic variants and QTLs on SSC 1, 8, 9, 13 and 18 for RFI and indicated significant biological processes and metabolic pathways involved in RFI. The study also detected novel QTLs for component traits of RFI. These results improve our knowledge of the genetic architecture and potential biological pathways underlying RFI; which would be useful for further investigations of key candidate genes for RFI and for development of biomarkers.
Figures

References
-
- Cai W, Casey DS, Dekkers JCM. Selection response and genetic parameters for residual feed intake in Yorkshire swine. J Anim Sci. 2008;86(2):287–298. - PubMed
-
- Kennedy B, Van der Werf J, Meuwissen T. Genetic and statistical properties of residual feed intake. J Anim Sci. 1993;71(12):3239–3250. - PubMed
-
- Gilbert H, Bidanel JP, Gruand J, Caritez JC, Billon Y, Guillouet P, Lagant H, Noblet J, Sellier P. Genetic parameters for residual feed intake in growing pigs, with emphasis on genetic relationships with carcass and meat quality traits. J Anim Sci. 2007;85(12):3182–3188. - PubMed
-
- Saintilan R, Mérour I, Brossard L, Tribout T, Dourmad JY, Sellier P, Bidanel J, van Milgen J, Gilbert H. Genetics of residual feed intake in growing pigs: relationships with production traits, and nitrogen and phosphorus excretion traits. J Anim Sci. 2013;91(6):2542–2554. - PubMed
-
- Muro-Reyes A, Gutierrez-Banuelos H, Diaz-Garcia LH, Gutierrez-Pina FJ, Escareno-Sanchez LM, Banuelos-Valenzuela R, Medina-Flores CA, Corral Luna A. Potential environmental benefits of residual feed intake as strategy to mitigate methane emissions in sheep. J Anim Vet Adv. 2011;10(12):1551–1556.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

