Replication of linkage at chromosome 20p13 and identification of suggestive sex-differential risk loci for autism spectrum disorder
- PMID: 24533643
- PMCID: PMC3942516
- DOI: 10.1186/2040-2392-5-13
Replication of linkage at chromosome 20p13 and identification of suggestive sex-differential risk loci for autism spectrum disorder
Abstract
Background: Autism spectrum disorders (ASDs) are male-biased and genetically heterogeneous. While sequencing of sporadic cases has identified de novo risk variants, the heritable genetic contribution and mechanisms driving the male bias are less understood. Here, we aimed to identify familial and sex-differential risk loci in the largest available, uniformly ascertained, densely genotyped sample of multiplex ASD families from the Autism Genetics Resource Exchange (AGRE), and to compare results with earlier findings from AGRE.
Methods: From a total sample of 1,008 multiplex families, we performed genome-wide, non-parametric linkage analysis in a discovery sample of 847 families, and separately on subsets of families with only male, affected children (male-only, MO) or with at least one female, affected child (female-containing, FC). Loci showing evidence for suggestive linkage (logarithm of odds ≥2.2) in this discovery sample, or in previous AGRE samples, were re-evaluated in an extension study utilizing all 1,008 available families. For regions with genome-wide significant linkage signal in the discovery stage, those families not included in the corresponding discovery sample were then evaluated for independent replication of linkage. Association testing of common single nucleotide polymorphisms (SNPs) was also performed within suggestive linkage regions.
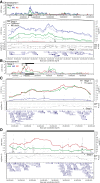
Results: We observed an independent replication of previously observed linkage at chromosome 20p13 (P < 0.01), while loci at 6q27 and 8q13.2 showed suggestive linkage in our extended sample. Suggestive sex-differential linkage was observed at 1p31.3 (MO), 8p21.2 (FC), and 8p12 (FC) in our discovery sample, and the MO signal at 1p31.3 was supported in our expanded sample. No sex-differential signals met replication criteria, and no common SNPs were significantly associated with ASD within any identified linkage regions.
Conclusions: With few exceptions, analyses of subsets of families from the AGRE cohort identify different risk loci, consistent with extreme locus heterogeneity in ASD. Large samples appear to yield more consistent results, and sex-stratified analyses facilitate the identification of sex-differential risk loci, suggesting that linkage analyses in large cohorts are useful for identifying heritable risk loci. Additional work, such as targeted re-sequencing, is needed to identify the specific variants within these loci that are responsible for increasing ASD risk.
Figures




References
-
- Autism and Developmental Disabilities Monitoring Network Surveillance Year 2008 Principal Investigators. Prevalence of autism spectrum disorders - autism and developmental disabilities monitoring network, 14 sites, United States, 2008. MMWR Surveill Summ. 2012;61:1–19. - PubMed
-
- Hallmayer J, Cleveland S, Torres A, Phillips J, Cohen B, Torigoe T, Miller J, Fedele A, Collins J, Smith K, Lotspeich L, Croen LA, Ozonoff S, Lajonchere C, Grether JK, Risch N. Genetic heritability and shared environmental factors among twin pairs with autism. Arch Gen Psychiatry. 2011;68:1095–1102. doi: 10.1001/archgenpsychiatry.2011.76. - DOI - PMC - PubMed
-
- Ozonoff S, Young GS, Carter A, Messinger D, Yirmiya N, Zwaigenbaum L, Bryson S, Carver LJ, Constantino JN, Dobkins K, Hutman T, Iverson JM, Landa R, Rogers SJ, Sigman M, Stone WL. Recurrence risk for autism spectrum disorders: a baby siblings research consortium study. Pediatrics. 2011;128:e488–e495. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

