NUP98-PHF23 is a chromatin-modifying oncoprotein that causes a wide array of leukemias sensitive to inhibition of PHD histone reader function
- PMID: 24535671
- PMCID: PMC4018760
- DOI: 10.1158/2159-8290.CD-13-0419
NUP98-PHF23 is a chromatin-modifying oncoprotein that causes a wide array of leukemias sensitive to inhibition of PHD histone reader function
Abstract
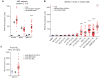
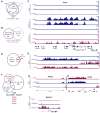
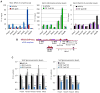
In this report, we show that expression of a NUP98-PHF23 (NP23) fusion, associated with acute myeloid leukemia (AML) in humans, leads to myeloid, erythroid, T-cell, and B-cell leukemia in mice. The leukemic and preleukemic tissues display a stem cell-like expression signature, including Hoxa, Hoxb, and Meis1 genes. The PHF23 plant homeodomain (PHD) motif is known to bind to H3K4me3 residues, and chromatin immunoprecipitation experiments demonstrated that the NP23 protein binds to chromatin at a specific subset of H3K4me3 sites, including at Hoxa, Hoxb, and Meis1. Treatment of NP23 cells with disulfiram, which inhibits the binding of PHD motifs to H3K4me3, rapidly and selectively killed NP23-expressing myeloblasts; cell death was preceded by decreased expression of Hoxa, Hoxb, and Meis1. Furthermore, AML driven by a related fusion gene, NUP98-JARID1A (NJL), was also sensitive to disulfiram. Thus, the NP23 mouse provides a platform to evaluate compounds that disrupt binding of oncogenic PHD proteins to H3K4me3.
Conflict of interest statement
Figures




References
-
- Hollink IH, van den Heuvel-Eibrink MM, Arentsen-Peters ST, Pratcorona M, Abbas S, Kuipers JE, et al. NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood. 2011;118:3645–56. - PubMed
-
- Pineault N, Buske C, Feuring-Buske M, Abramovich C, Rosten P, Hogge DE, et al. Induction of acute myeloid leukemia in mice by the human leukemia-specific fusion gene NUP98-HOXD13 in concert with Meis1. Blood. 2003;101:4529–38. - PubMed
-
- Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol. 2007;9:804–12. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

